 I think we’re focusing on the wrong thing when we follow classical textbook approaches to ‘Biomolecules’ and their categories (DNA, RNA, protein, carbs, lipids). I don’t have much to say about the latter two, but there are some Big Ideas about the molecules of the central dogma that I believe get lost in the tedium of -OH groups and dehydration reactions… I’ll discuss order of presentation and such later; now I want to talk about monomer structure. To wit: How are (the group nucleic acid monomers) and (the group amino acids) all similar, and why must this be so?
I think we’re focusing on the wrong thing when we follow classical textbook approaches to ‘Biomolecules’ and their categories (DNA, RNA, protein, carbs, lipids). I don’t have much to say about the latter two, but there are some Big Ideas about the molecules of the central dogma that I believe get lost in the tedium of -OH groups and dehydration reactions… I’ll discuss order of presentation and such later; now I want to talk about monomer structure. To wit: How are (the group nucleic acid monomers) and (the group amino acids) all similar, and why must this be so?
Short form: the molecules of the Central Dogma (DNA, RNA, proteins) are special; creating strings of these requires that a single machine (DNA and RNA polymerases; the ribosome) join strings of any sequence, whereas most enzymes perform single, stereotypical reactions. What must we do to allow DNA Pol to put together ANY pair of nucleotides, and the ribosome to add ANY amino acid to another?
Note: This post focuses on the commonalities of the structures–how they are joined; how single machines put a diversity of pieces together. For an overview on the central dogma itself, try this post; or specifics on DNA or amino acids or translation.
I’m a strong believer that sensible things can be taught to students “even though they’re freshmen”. If you think about it, the only reason they’re likely to be ‘smarter’ as upperclassmen is that we’ve contributed to making them that way. So shouldn’t we start early and be active about it?
In looking at the ‘parts’ of an amino acid, it’s easy to get caught up in the Naming of Parts –alpha carbon, amino group, carboxyl group… but why? I think the answer’s simple and obvious, in hindsight: if the ribosome had to have an active site for every combination of amino acids it might need to join together, that’d be 400 active sites!
What to do instead? Easy! Have each unit contain a stereotypical part on which the ‘sewing machine’ (polymerase/ribosome) operates, and a ‘business end’ that confers its unique operational properties. Since all 20 amino acids have ye amino and carboxyl groups, identical recognition and chemistry events can be performed on all, regardless of order, regardless of properties.
The same core idea applies to nucleotides. No matter which is being joined to which, the chemistry is nucleophilic attack by 3’OH of a ribose (or deoxyribose–not directly relevant here) to the inner phosphorous atom of a triphosphate attached to the 5′ position of an incoming nucleotide.
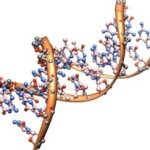 DNA vs. RNA: Go Big or Go Home
DNA vs. RNA: Go Big or Go Home
I think halfway measures are the death of constructivism and introducing Big Ideas to introductory students. The differences between DNA and RNA are critical and readily understood… if we explain them in discussing the molecules of the central dogma. On the other hand, if they are rendered only as lists of compare-and-contrast, then we are adding meaningless terms as sub definitions of… other meaningless terms. If called upon, I could cheerfully defend the position that at the introductory level, there’s nothing critically different between DNA and RNA. Both…
- Are joined via 3′-OH attacks on 5′-triphosphates
- Have a base-sugar-phosphate structure
- contain ‘information’ in terms of specific sequences that can be complemented and thereby copied
- can serve as the template for each other (DNA => RNA is RNA Polymerase; RNA => DNA is reverse transcriptase)
- mediate partner-selection via basepairing
Their differences generally don’t feature in any discussion of introductory biology (though both are fantastic evolutionary discoveries whereby a superior product [DNA] displaces its antecedent [RNA]; an explanation of thymine vs. uracil here, and its lecture materials here). Condensed form:
- thymine and uracil are both partners of adenine
- thymine differs from uracil at a non-basepairing position
- recognition of the difference (methyl group) on thymine generally doesn’t feature in Introductory Biology discussions
- the absence of the 2′ -OH in deoxyribose is all about preventing something from happening (attack of the 2′ -OH on the adjacent 3′ -OH linkage to the rest of the chain; a successful attack breaks the strand). Again, if this conversation doesn’t take place, the difference between DNA and RNA here is arbitrary.
On the other hand, my preferred approach is to attack these interesting (and understandable) differences by devoting a couple class periods to investigating them. The uracil vs. thymine story offers deep insights into how evolution works, possibly into the origins of life, and challenges students to think ‘like a cell.’. The advantage of eliminating the 2′ -OH on RNA reinforces the structure of nucleic acid chains and ties in to the Sn2 reactions (this link goes to a discussion of ATPase mechanism, of which such a reaction is part), which I have introduced in my own class by the time DNA/RNA structure is covered.
Nucleic acid vs. protein: instructions vs. machinery
I think the Big Idea we need to be communicating to students here is functional aspects of structure of the molecules of the central dogma. Teaching them that the ‘Genetic material’ uses ‘bases’ or ‘nucleotides’ which have ‘ribose’ and are synthesized “5′ to 3” while ‘proteins’ are made of ‘amino acids’ which have ‘carboxy termini’ and are synthesized “N-to-C” adds words upon words, with no clear requirement that any of them be anchored in concepts. The critical differences and the critical features all tie in to structure. Nucleic acids have the special ability of partnering specifically and unambiguously. Thus they can be copied and their sequences maintained. Amino acids represent a toolkit filled with diverse ‘feels’ (interaction capabilities realized through neutrality, positive & negative charge, or H-bond donor and acceptor). Nucleic acids hold instructions; amino acids are building blocks for machines. These ideas are also discussed here.
Making it an exploration: ‘Info Flow’
thinkBio’s ‘Info Flow’ is an exercise designed to capture both the roles and the features-that-fit molecules to roles. As of January 2015 it’s still getting revised with new thinking and adaptation to lessons learned in offering it in classrooms, but versions can be explored here.
Further explorations
More on the ‘matching’ of molecules to functions in the Central Dogma
How amino acids are well-fitted to the task of ‘building blocks of cellular machines’
How self-assembly is a critical–and common–feature of DNA, RNA, proteins, membrane components
Thinking about… and teaching… the structures of biological molecules
Rant: condensation reactions
We seem to get excited about the fact that according to TEXTBOOK DRAWINGS, adding nucleotides or amino acids together ‘pops out’ a water. This is indeed the NET chemistry, but in neither case is it what happens in detail. When adding an NTP to an available 3′ OH group, the leaving group is actually PPi (inorganic diphosphate), NOT HOH. Similarly, since amino acids are added after delivery by tRNAs (to which they are covalently attached), the leaving group is tRNA-OH, not H-OH.
I think the problem is the lure of objective, easily graded questions. These are ‘factual’, and not as conceptually challenging as the above. But in my opinion, they’re empty calories. Would life be different if the leaving group were NH3 instead of H2O? Not that I know of. What matters is that all the backbone elements are the same… not the details of that sameness.
Pingback: What's important about... Concepts in Translation
Pingback: Teaching basepairing: consequences, applications
Pingback: Hard choices: what NOT to teach in Introductory Biology
Pingback: Talk different: delivering biology with meaning
Pingback: Translation in biology: the Sense of Things