‘What to teach’ in Introductory Biology presents a whole host of conundrums. Is our role to cover the breadth of all of biology (or all of the biology student will hear again in upper divisions) at breakneck pace and without depth? A whirlwind tour of vocabulary to commit to memory with vague understanding for some later date? I will argue periodically that we should pick a limited, integrated fraction of possible content and teach it in a way that allows students to see and grasp underlying concepts and universal themes, thereby enabling them to figure out what comes later, and to embark on their own to investigate whatever catches their fancy.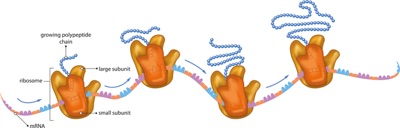
The key ideas of biology’s Central Dogma–or better put, the flow of information— are a critical case in point. We all agree that ‘something’ about DNA, RNA and protein is among our core duties. But what? My view is that identifying the roles of each of these players, how their structures fit them to play those roles, how they came to occupy and the ‘flow’ between them them are straightforward, core ideas in biology.
Overview
Biology’s central dogma is a funny beast. The phrase’s origin lies partly in the fact that Crick didn’t correctly understand the meaning of the term ‘dogma’ when he coined it (by his own admission). More to the point: what is the essence here? Every textbook and every Intro Bio lecture covers something about DNA, about RNA, and about proteins. DNA replication, transcription and translation are guaranteed to feature. Textbooks tend to focus exclusively on “These are the chemical structures. These are the steps of the processes”. When we follow their lead, I think students are left confused and lost–one misconception we’ve seen at my University is that students think that in order to produce a protein, cells first undergo DNA replication, then transcribe the entire genome, then proceed to translation. It seems funny, but when you look at the order of chapters and the fact that we often fail to talk about purpose to them, it’s not all that far-fetched.
Note: these ideas are encapsulated in an introductory way in two software explorations at thinkBio: “InfoFlow DNA” and “InfoFlow Protein”; both are found here (alas, the Mac versions no longer works; I’m not sure about the Windows ones. Briefly, they were an effort to conceptualize and analogize the flow of information via molecules and interactions).
I want to take a wholly different approach. I’m going to suggest that the keys to the kingdom lie in asking what is the purpose of each molecule, and how do the structures of its building blocks align each to its role. As always, I’m not a fan of the catch-phrases that make for such wonderful multiple choice selections such as “information molecule of the cell”. Information in awfully hard to pin down; try to verbalize a quick, sound definition… and then see how you apply it to cells. I have come to strongly favor the term ‘instructions’. Students ‘get’ instructions; after all, we spend a lot of class and exam time foisting such on them. More importantly, ‘instructions’ aligns very well with the biological role of DNA. My favorite definition of a gene is “The instructions for how, when, where and how much to make of a specific product, often a string of amino acids.” Handily, this captures regulation as well as coding sequence.
A bit of preamble
In a conversation my colleague, Emily Dykstra, challenged me to come up with a succinct, conceptual level description of biology’s Central Dogma vis-a-vis teaching Introductory Biology. Here are two passes I took at it:
Life requires the ability to instruct the creation of machines; these instructions must be controlled and passed to offspring. ‘Instruction molecules’ must be ’self-copyable’ (based on ‘automatic chemistry’)*. The molecules that perform the actions must arise through mechanical or automatic processes directly from the instructions.
*By ‘automatic chemistry’, I’m trying to point out that basepairing needs no ‘paired’; it is the chemical nature of the bases to stabilize as pairs (of course, in competition with water pairing and Brownian Motion, a single pair doesn’t count for much)
An alternate presentation:
We need instructions and a way for the instructions to be turned into actions. The instructions must be capable of ‘having evolved’ and must be copy-able, both for short-term use as well as inheritance. Actions arise from actors, and there must be a ‘boring’ process by which a near-infinite number of functional actors can be created using finite machinery from the instructions.
Overall: functions of life must arise directly from chemistry, and the chemistry of the different classes of molecules will directly reflect their roles.
Biology’s Central Dogma: Order of Presentation
Before going on, I’d like to fire a shot across the bow of teaching sequence for these concepts. Given that instructions are pretty nebulous until a conversation regarding instructions for what has taken place, I strongly question the classical order-of-introduction, which follows the arrows in the central dogma (DNA first, then RNA, then protein). It seems to me that proteins are the most readily understood members of the Big Three–they are machines that do stuff. Everyone can relate to machines; by choosing some concrete examples, we can have students heads in the molecular game. My favorite (and that of thousands of others!) is hemoglobin (this link takes you to a discussion of hemoglobin as an organizing theme for teaching many Intro Bio concepts). By starting with a given protein and understanding the role of selected amino acids in its function, I don’t end up in the awkward position of talking about the instructions without having a concrete answer to the question ‘instructions… for what ?‘
Proteins
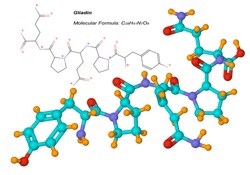 I’ll start with the case of proteins and their role. I commonly ask students to think about amino acids as construction kits. There is a very simple question: if you were locked in a room with a set of Legos, which would you rather have: a kit with 2-3 different pieces, or one with dozens in a delightful variety of shapes, sizes, and perhaps even functionality? From there, asking them to predict what the cell’s tool-constructing kit looks like is trivial: they predict that it should have a lot of diversity to it. Having already covered (in class, though not yet in this blog) the fact that there are only a small number of ‘ways to be’ chemically (positively or negatively charged; neutral, H-bond donor or acceptor), we expect to find representatives of every type here. And do! Further, we can class amino acids by size, flexibility, etc. and in every case we find diversity. For a discussion of how the common structural elements uniting all amino acids come into play, go here.
I’ll start with the case of proteins and their role. I commonly ask students to think about amino acids as construction kits. There is a very simple question: if you were locked in a room with a set of Legos, which would you rather have: a kit with 2-3 different pieces, or one with dozens in a delightful variety of shapes, sizes, and perhaps even functionality? From there, asking them to predict what the cell’s tool-constructing kit looks like is trivial: they predict that it should have a lot of diversity to it. Having already covered (in class, though not yet in this blog) the fact that there are only a small number of ‘ways to be’ chemically (positively or negatively charged; neutral, H-bond donor or acceptor), we expect to find representatives of every type here. And do! Further, we can class amino acids by size, flexibility, etc. and in every case we find diversity. For a discussion of how the common structural elements uniting all amino acids come into play, go here.
Not only are the amino acids diverse in these ways, we can also identify ‘specialists’ in the collection. Glycine essentially has no side chain, making it a wiz for tight corners. Cysteines are capable of ‘easily’ forming covalent bonds, allowing for sturdier proteins ‘locked’ into desired configurations. Histidines are sensitive to pH in biologically relevant ranges (technical: pKa is about 6.5), and indeed they are often the cause of pH responsiveness in proteins (again, hemoglobin; see here for the totally cool, pH-sensitive miraculin protein)[FYI, miraculin pills, available from Amazon and thinkgeek.com are a really fun way to introduce pH concepts in class]
DNA
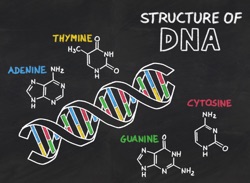 Having established what-to-build, let’s consider the nature of the instructions themselves. Our expectations are now completely different. What do we want for a set of copyable instructions? The majority of the relevant ideas are covered in my post on DNA; the short form is we want inflexible, weakly interacting, unambiguous pieces that match one and only one partner (it can be self; our code is complementary instead). We expect (and get) a small number of pieces because we’re looking for accurate matching here–the fewer pieces we have, the more different they can be from one another, and the smaller the likelihood that two non-partners will ‘find a way’ to pair up.
Having established what-to-build, let’s consider the nature of the instructions themselves. Our expectations are now completely different. What do we want for a set of copyable instructions? The majority of the relevant ideas are covered in my post on DNA; the short form is we want inflexible, weakly interacting, unambiguous pieces that match one and only one partner (it can be self; our code is complementary instead). We expect (and get) a small number of pieces because we’re looking for accurate matching here–the fewer pieces we have, the more different they can be from one another, and the smaller the likelihood that two non-partners will ‘find a way’ to pair up.
More: thinkBio eBook chapter on DNA & RNA
RNA
 RNA, the ‘man in the middle’, is basically a cheap knock-off of DNA… except that such a view tells the tale backwards. Evolutionarily speaking, RNA is a flawed “Mark I” version of a genetic material… but one with the adorable property of being able to fold into complex 3D structures (like proteins) that can act as enzymes… like proteins. Sure, it has shortcomings–the -OH on the 2′ ribose position make it vulnerable to self-destruction, and its use of uracil render inevitable chemical changes to cytosine damaging and undetectable, but its current niche is that of short-term copy. In this role, disposability is an advantage (many of the enzymes that destroy RNA simply facilitate the self-destruct pathway available through the 2′ -OH group).
RNA, the ‘man in the middle’, is basically a cheap knock-off of DNA… except that such a view tells the tale backwards. Evolutionarily speaking, RNA is a flawed “Mark I” version of a genetic material… but one with the adorable property of being able to fold into complex 3D structures (like proteins) that can act as enzymes… like proteins. Sure, it has shortcomings–the -OH on the 2′ ribose position make it vulnerable to self-destruction, and its use of uracil render inevitable chemical changes to cytosine damaging and undetectable, but its current niche is that of short-term copy. In this role, disposability is an advantage (many of the enzymes that destroy RNA simply facilitate the self-destruct pathway available through the 2′ -OH group).
By teaching students the roles each of these molecules occupies, we can lead them to good predictions about their properties. From that position, an examination of their structures makes sense, which seems the best foundation for teaching.
Further explorations
How amino acids are well-fitted to the task of ‘building blocks of cellular machines’
Building the polymers: pieces are built so that the strings (polymers) can be made by single machines
Self-assembly is a critical–and common–feature of DNA, RNA, proteins, membrane components
Thinking about… and teaching… the structures of biological molecules