 Everybody agrees that we should teach the ‘Central Dogma’ in IntroBio (DNA => RNA => Protein). But surely that’s not an answer to the question. What is important to understand about the actors and the ‘flow of information’ [what’s information?], why does each actor have the role it has, why are the actors chemically as they are and not otherwise (to paraphrase Einstein). As part of this blog, I’m going to go through views I’ve developed over two decades, starting with DNA. Foreshadowing: it’s not going to be about alphabets: the letters D-N-A and the letters A-G-C-T cannot encompass what is important about a molecule capable of holding in its structure the instructions-to-build for every known living thing.
Everybody agrees that we should teach the ‘Central Dogma’ in IntroBio (DNA => RNA => Protein). But surely that’s not an answer to the question. What is important to understand about the actors and the ‘flow of information’ [what’s information?], why does each actor have the role it has, why are the actors chemically as they are and not otherwise (to paraphrase Einstein). As part of this blog, I’m going to go through views I’ve developed over two decades, starting with DNA. Foreshadowing: it’s not going to be about alphabets: the letters D-N-A and the letters A-G-C-T cannot encompass what is important about a molecule capable of holding in its structure the instructions-to-build for every known living thing.
My central argument is going to be that in teaching DNA, the essence of nucleic acids is copy-ability. It is this feature that lead Crick and Watson, writing up the solved structure, to famously comment
What, then, are specific features of bases that contribute to ‘specific pairing’? What are we looking for? In introducing these ideas to students, I use a short classroom exercise. I first ask students to shake hands with at least 3 others in the classroom. I then ask how many failed. I’m often met with quizzical looks; what kind of idiot could fail to shake hands any number of times? That’s the set-up: I then ask them to close their eyes and imagine having their hand shaken by 10 people. How many think they could spot the 3 the first shook with?
Conclusion: the ‘regular’ handshake contains no ‘information’ about the partner; there is no ‘specificity’.
Next, the class is paired off and each group is tasked with developing a ‘secret handshake’ that, if surreptitiously offered, could be used to recognize fellow members of the club. I stipulate that this handshake cannot involve motion (I got a lot of ‘bird flapping’ gestures the first time :-). We then take a quick look at several volunteers. Examples include the ‘Spock live long and prosper‘ structure, with one partner offering the hand parallel to the ground, and one perpendicular, with the ‘V’ connecting.
I ask them to find features important to their secret handshake; with prompting, I’m generally able to get ‘rigid, structurally specific’. Intriguingly, most tend to be complementary, though this isn’t inherent to the exercise.
I then go for one more element that many overlook by inquiring whether Shaquille O’Neal could shake their hand if he decided that’s what he wanted to do. Everybody pretty much agrees he could–in other words, a STRONG partner can defeat SPECIFICITY: weakness is accuracy.
This then forms the basis for PREDICTIONS about properties we expect in the storage/instruction molecule of the cell:
- it will be structurally rigid
- there will be clearly defined interactions
- correct ‘fit’ will be easily detected
- interactions will be multiple WEAK ones rather than mighty ones that can each ‘go it on their own’.
Molecular Answers
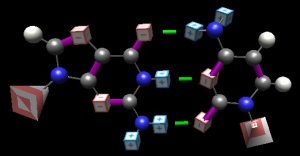
A GC pair, rendered in ‘BasePairer‘ (old form). Partial positive charges shown is blue cubes; partial negative as red. The ribose-phosphate groups are shown as pyramids; their orientation reminds you of the anti-parallel nature of DNA. Hydrogen bonds indicated as green.
The nucleotide bases do a lovely job of filling this prediction. The ring structures, filled as they are with alternating double-and-single bonds, are pancake-flat. There are no alternating 3D structures (see ‘chair and boat‘ conformations of ring sugars, for example)
There is also rigidity of presentation of the hydrogen bonding (hence H-bonding) positions: the double-bonded oxygens can go nowhere, the hydrogens in the central position are equally constrained; only the nitrogens in the -NH2 groups can move–and all they can do is rotate about the bond to the ring structure.
Hydrogen bonding is a ‘binary code’: one is either a donor or an acceptor, and only opposites attract. Thus there is potentially greater ‘meaning’ than in hydrophobic ‘interactions’, for example, where any one hydrophobic surface ‘goes with’ any other.
Weakness is often overlooked, but is a critical element, analogous to the O’Neal example. Hydrogen bonds are not only small of contribution (in a water environment, that is), but they are very limited in their abilities–only at a narrow range of differences, and best when the three atoms involved (the Hydrogen, the O or N atom to which it is covalently bonded, and the O or N to which the weak interaction forms) are in a linear array.
The bases thus provide everything one could wish for in terms of specific partnering… which is at the heart of ‘copy-ability’.
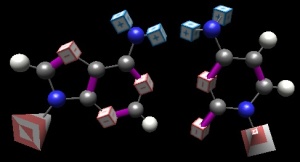
Non-pairing: just as important as pairing!
In teaching DNA, an often (always?) overlooked aspect of our genetic code is the horrible ‘fit’ between the non-pairs: A:C and G:T. However, it is inescapable that this failure-to-pair is just as important as the focused-upon ability of Gua and Cyt, Ade and Thy to pair. If one of the bases could, at random, interact with either of two partners, information would be lost at every round. A quick examination of these non pairs (BasePairer) reveals that the 4 ‘code members’ we have a great at failing to pair (the interesting case of the skewed ‘discovery’ of two H-bonds between G and T, which is common in RNA structures, must await another day).
Rant: Anti-parallel: Big or Little deal?
We make a big deal about making students memorize that DNA ‘goes’ 5′ => 3′ and that the strands are ‘anti-parallel’. What have they understood when they memorize this? Do we do a good job of conveying “the 5′ of what”? Perhaps most importantly of all, why does this matter? If we cannot tell them how life would be different if the two strands of DNA are parallel, and they don’t really understand what it means structurally that they’re ‘anti-parallel’, what is being taught and learned?
FYI, in my opinion, there is a very good reason why we ended up with components that settle into an antiparallel rather than a parallel structure. I’ll bet money that if an effort were made to discover catalytic nucleotides using a parallel-strand-forming structure, they’d be few and far between.
Why? Try this simple experiment. Draw the classic, 2-D ‘cloverleaf’ structure for a tRNA, noting all the regions of double helix. If drawn correctly, your 5′ and 3′ ends close together at the ‘top’
Now: Try to draw the same structure but requiring the base pairing strands to be PARALLEL. For my money, that’s why the ‘winning’ structure here on Earth (and those elsewhere) was parallel.
What’s next
Related to this piece, I intend to write
–Perfect partnering in therapeutics: gene re-writing (CRISPR); gene inactivation (siRNA) [May appear as lecture module]
–Proofreading: how a stupid machine can check its own work
–Mutation: How the chemical nature of the code combines with partnering to make error inevitable
Pingback: What's important about... Key Concepts in Biology
Pingback: Teaching basepairing: consequences and applications
Pingback: Hard choices: what NOT to teach in Introductory Biology
Pingback: What's important about... molecules of the Central Dogma?
Pingback: A mechanistic view of the cell: the glory of because