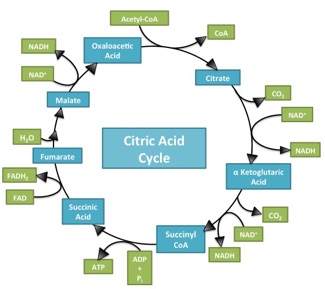
What makes the Citric Acid Cycle (Kreb’s cycle, TCA cycle) ‘hard’? Misdirection. When most of us sit down to learn something, the first question is usually “what is important here”. When trying to figure out a complicated process (Americans: think about the sport of cricket!), the questions are “What should I be looking at? And what am I seeing when I look there?”. It’s deeply ironic, then, that in looking at Introductory Biology textbooks, it’s incredibly difficult to spot an electron anywhere in representations of the citric acid cycle. This is bizarre, in that the cycle has only two jobs: 1) extract ‘high value’ (energetic) electrons, and 2) be a cycle so it can keep… cycling. A call for electron-watching and some suggestions for teaching electron flow through the citric acid cycle follow (Image source) Continue reading

 using different insertions for the variables”, the game gets much more complicated… and there’s a lot more to ‘teach’ and to discover. I’ve posted a number of different “derived wisdoms” elsewhere (
using different insertions for the variables”, the game gets much more complicated… and there’s a lot more to ‘teach’ and to discover. I’ve posted a number of different “derived wisdoms” elsewhere ( Too often, biology is presented as gathering data and letting it lead us. While this is a valid and important component, there are also many glorious examples where the thinking came first and directed the experimentation. Many of the great experiments in biology came about this way–Meselson and Stahl (semi-conservative replication), Hershey-Chase (the Blender experiment, or ‘who is the genetic material)… and some that are more abstract–Pauling’s proposal of the alpha helix and beta sheet… and a number of Crick’s hypotheses. This post covers his prediction that efficiencies would be achieved in the tRNA pool through wobble pairing: relaxing orientation of the participants in a basepair to allow one base to partner with several others.
Too often, biology is presented as gathering data and letting it lead us. While this is a valid and important component, there are also many glorious examples where the thinking came first and directed the experimentation. Many of the great experiments in biology came about this way–Meselson and Stahl (semi-conservative replication), Hershey-Chase (the Blender experiment, or ‘who is the genetic material)… and some that are more abstract–Pauling’s proposal of the alpha helix and beta sheet… and a number of Crick’s hypotheses. This post covers his prediction that efficiencies would be achieved in the tRNA pool through wobble pairing: relaxing orientation of the participants in a basepair to allow one base to partner with several others.