Too often, biology is presented as gathering data and letting it lead us. While this is a valid and important component, there are also many glorious examples where the thinking came first and directed the experimentation. Many of the great experiments in biology came about this way–Meselson and Stahl (semi-conservative replication), Hershey-Chase (the Blender experiment, or ‘who is the genetic material)… and some that are more abstract–Pauling’s proposal of the alpha helix and beta sheet… and a number of Crick’s hypotheses. This post covers his prediction that efficiencies would be achieved in the tRNA pool through wobble pairing: relaxing orientation of the participants in a basepair to allow one base to partner with several others.
Too often, biology is presented as gathering data and letting it lead us. While this is a valid and important component, there are also many glorious examples where the thinking came first and directed the experimentation. Many of the great experiments in biology came about this way–Meselson and Stahl (semi-conservative replication), Hershey-Chase (the Blender experiment, or ‘who is the genetic material)… and some that are more abstract–Pauling’s proposal of the alpha helix and beta sheet… and a number of Crick’s hypotheses. This post covers his prediction that efficiencies would be achieved in the tRNA pool through wobble pairing: relaxing orientation of the participants in a basepair to allow one base to partner with several others.
Background: the pattern
As the ‘codon table’ (which mRNA nucleotide triplets ‘coded for’ which amino acids) were being filled in, a number of patterns became evident (in the figure below, the first ‘letter’ of the codon is in the center, then select outward–second codon, third codon).
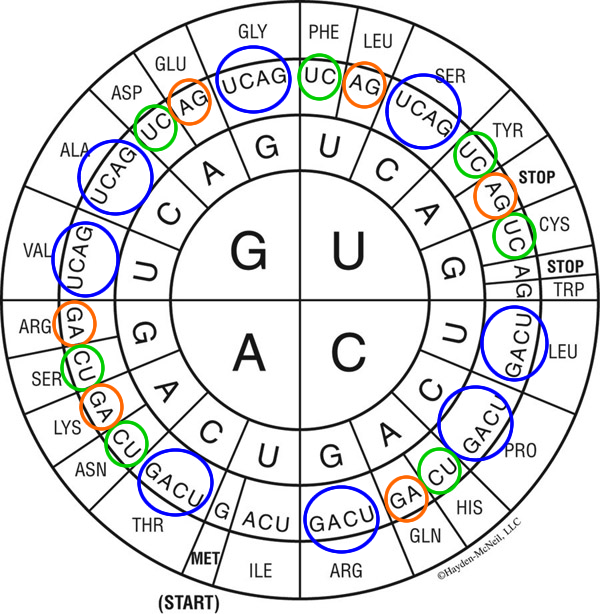
Notably, in many cases, all four possible ‘third nucleotide’ position selections ‘call for’ the same amino acid–at the top of the figure, going from the central ‘G’ upwards, note that G-G-[UCAG] all code for glycine. This pattern holds for eight of the amino acids (blue circles). For seven, either purine (A, G) results in the same outcome (orange circles) and for another seven, either pyrimidine (C, U) calls for one amino acid (green circles). Lastly, for one amino acid (isoleucine; just left of bottom) A, C and U all ‘answer’ if the codon starts A-U-.
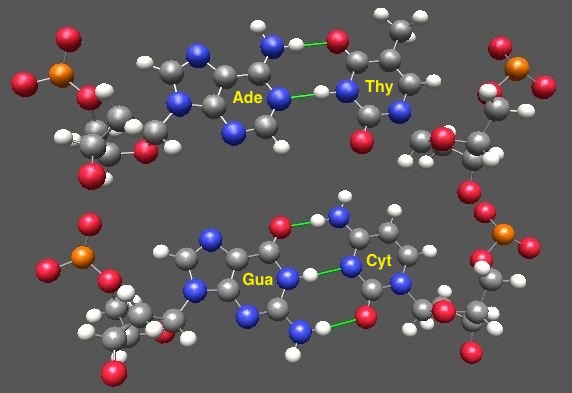
Word reminder: the base is the part that does the actual pairing via hydrogen-bonding interactions; once it’s attached to the sugar and phosphate, the whole thing is called a nucleotide. So a base is a subset of a nucleotide, and retains its specific partnering capability. The classical basepairs, guanine-with-cytosine and adenine-with-uracil, are shown below (green lines mark the non-covalent hydrogen bonds)
Process: identifying what’s ‘noteworthy’ (salient)
What to make of this? One reasonable answer is that if several similar codons ‘mean’ the same thing, then errors will be less likely; an ‘oops’ in matching at that position won’t have consequences. But Crick was a very thoughtful dude. He was accustomed to thinking about basepairing (after all, combined with Franklin’s crystal data, it was the matching of the basepairing positions that lead to the proposed structure of DNA!). It occurred to him that there were possible efficiencies here, in that for the tRNA anticodons (the 3 nucleotides that basepair with the codon) two of the bases would be identical for codons that themselves shared two letters (i.e. if all glycine codons start “GG-” then all glycine tRNA anticodons must end “-CC” (remember, pairing in DNA and RNA is anti parallel).
Crick also knew that evolution was a great experimentalist… and often ‘finds a way‘ (with both anger & apologies to Jurassic Park). Is there a way that a single tRNA could handle two, three or even four codons? Well, yes, yes, and no–at least that’s what’s going on in your cells right now. Let’s work backwards from the answers to the evidence… starting first with why this better be the exception to the rule.
As I’ve presented elsewhere, the genetic material (yes, the same bases we’re talking about here!) has one job–specific basepairing (no ambiguities! single partners for everyone!). For this task specifically, the ‘best’ bases to use would have no hope of ‘discovering’ ways to fit together. But with only a ‘binary’ interaction code (the partial (+) of hydrogen bond donors and the partial (-) of hydrogen bond acceptors) that’s going to be a hard goal to hit. Further specificity comes from the fact that the double helix itself, as well as the machines charged with doing copying (DNA and RNA polymerase) require the bases do their matching under tight positioning constraints–the ribose sugar must be ‘just so’… and since the ribose is covalently attached to the base, it’s not going anywhere either. But positioning during translation is a horse of a different color. Here, the ‘double helix’ is only three basepairs long (the codon of the mRNA and its partnership with the tRNA anticodon). So at each end of the helix, there are no neighbors ‘demanding’ that the adjacent base/ribose ‘straighten up and fly right’. Similarly, the region of the ribosome that constrains the positioning of the bases is whatever it has evolved to be. If the pocket was tight fitting, then the bases would have to be wherever they were ‘told’ to be. But if the pocket on the ribosome allowed one of the end bases to get a little loosey-goosey (say, the 1st [5′] base of the tRNA anticodon) then what partnerships might be discovered?
Discovery: basepairing exercises
Teaching moment: the best learning is always discovery learning. Do you want to explore for yourself what a guanine could do with a thymine, if given a little freedom? Remember, for many of the codons, both cytosine and uracil in the 3rd position ‘mean’ the same amino acid; if guanine can ‘find a way’ to interact with uracil, a single tRNA with guanine could pair with either ‘flavor’ of codon. To examine this possibility, take a look at guanine + uracil.
But things get a good deal fancier than this! One of the noteworthy things about tRNAs is that they include significant numbers of non-(A,G,C,U) nucleotides. While they’re transcribed ‘as normal’ from DNA by an RNA polymerase, afterwards several exotic things are done to them enzymatically. One of these is that some have their 5′ adenine modified to inosine (remember, the 5′ anticodon position pairs with or ‘reads’ the 3′ or 3rd nucleotide of the codon). Why? Well, again recalling that 2 hydrogen bonds is sufficient to make a pair (after all, that’s all A-U/A-T make!), go exploring here and see if you can pair inosine with A, C and U!
Thought experiment: Crick ‘sees’ the code
Of course, at the beginning of all this, I made the claim that Francis Crick thought all this up prior to the data. Notably, he did it doing essentially what you did in the links above. Here’s Crick’s paper outlining his thinking and figures supporting it. Pay particular attention to Fig. 6, the actual ‘thought experiment’; while it’s a bit of tough going, here are some guides: 1) in all cases, Crick is representing a particular carbon atom from the ribose sugar so all the pairs are conceptually ‘overlaid’. 1) in all cases, he holds this atom from the anticodon constant; it’s marked with X. 1) In the case of the ‘normal’ basepairs, the corresponding carbon on the partner is in the exact same position (all basepairs ‘look alike) 3) the off-kilter relatives positioning of the ‘wobble’ partnering are indicated by all the other ball-and-stick positions. You can easily see how each differs from the ‘standard’ positioning that is required by the DNA helix, and by DNA and RNA polymerases. That’s how much freedom the ribosome allows in wobbling! And that’s how one tRNA can ‘service’ multiple codons.
Lastly, here are the ‘pairing answers in the back of the book’ at Wikipedia… though if you made the requisite number of hydrogen bonds, you should already know you’re right!
You might also enjoy: further explorations