Too often we overlook the tough job that tRNAs do, seeing them only as the handmaidens of translation: matching up with mRNA; delivering a specific amino acid place on their heads by synthetases, arriving at the ribosome to first take the chain and then be stripped of it… These tiny (~100 nte.) elves perform a series of amazing sleights-of-hand: sometimes they must all be the same (the ribosome must deal similarly with all of them); at other times, unique (codon matching) and still other times, as ‘family members’ (all those ‘serviced’ by a single tRNA synthetase. Just how much recognition-surface and information is stamped onto a tRNA?
Credit where due: this post began as a conversation with Kait McLeod, who lauded the many fascinations of protein in a conversation we were having, forcing me to stand up for RNA; I selected tRNAs as the weapon of my argument. There’s a great internet meme out there: “You had only one job…”. That’s not tRNAs. Per Shakespeare, each actor in its time must play many roles. What’s most striking (to me) is that sometimes it’s critical for the whole group of them to be “all alike”, whereas at other times, they must be ‘each unique’. As is the case with other biological molecules, this is achieved by a common chassis covered with unique ‘decorations’ (amino acids: backbone common; sidechain unique; nucleotides ribose-phosphate common; base unique; lipids glycerol common; fatty acids distinct). With tRNAs, the primary distinction is STRUCTURE common; specific bases unique (including ‘enhancements’–covalent modifications to bases such that tRNAs sport far more than the canonical Big 4, though they’re each synthesized as ‘classical’ RNA copies of a DNA strand).
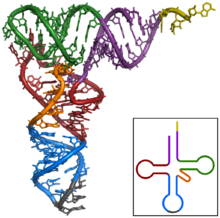
tRNA: the amino acid adds on the yellow extension at top right; the anticodon is gray at bottom
Source: https://upload.wikimedia.org/wikipedia/commons/thumb/b/ba/TRNA-Phe_yeast_1ehz.png/220px-TRNA-Phe_yeast_1ehz.png
Organizing this is challenging; I have opted to go from the ‘all the same’ classes to the ‘each unique’ classes as the former is ‘big’ and the latter smaller.
They’re all the same!
tRNA structures all the same: for the ribosome
As I’ve presented before, one recurrent theme in molecular biology is the ‘common chassis/unique attachment‘ pattern. It holds in tRNAs in a rather unique implementation–from a distance, tRNAs are the same size (about 100 nte.) and shape (the ‘cloverleaf’ 2D structure in the image at lower right; the folder ‘inverted L’ for the 3D structure in the primary image). This matters. As discussed previously, you can’t have a unique machine to add each different component–you get an infinite number of builders-of-builders built by builders-of-builders-of-builders built by… Instead, you need generic assembly steps completed by machines that can act on any of the pieces… then the strings-of-pieces must ‘automagically’ form themselves in 3D.
This means the ribosome must be able to ‘handle’ any tRNA and its amino acid. The ribosome can’t very well have 20+ active sites; instead the burden falls on the tRNAs to all ‘fit’ into the ribosome and to each be recognized by and acted on by the ribosome. Each tRNA must ‘sit’ such that when its anticodon touches the codon, the tRNA ‘tickles’ the ribosome to ‘realize’ that everything is good. When in position, the tRNA must ‘present’ its amino acid cargo in exactly the right place to receive the currently constructed peptide chain.
tRNA structure all the same: EF-Tu
We don’t talk about it much in Intro Bio, but the tRNA+amino acid combo alone is not sufficient to join the dance of translation. There’s a critical accompanying protein factor, name EF-Tu in E. coli (‘Elongation Factor’). This GTP-hydrolyzing machine is a critical component of accuracy in translation. Since every tRNA must bring one to the ribosome, every tRNA must ‘look alike’ to EF-Tu in order to gain its favor (binding)
Interlude: tRNA structures each different: EF-Tu part II (advanced)
Interestingly, the story is more subtle than just “hey, let’s all go out for an EF-Tu!”. This is a bit of an advanced topic, but Uhlenbeck and colleagues (Ref#1, Ref#2) have shown that the ‘stickiness’ of EF-Tu for individual (tRNA + amino acid) pairs is ‘tuned’ to further enhance accuracy. Briefly, the sum (stickiness of EF-Tu for amino acid cargo) and (stickiness of EF-Tu for ‘top’ of tRNA) has evolved to roughly a constant, with EF-Tu ‘liking’ big, hydrophobic amino acids ‘pretty much’ and their tRNAs less so. What’s the point? It’s incredibly elegant: if a tRNA synthetase (the machines that put tRNAs together with their hopefully correct amino acids) makes a mistake, then the EF-Tu ‘stickiness’ for the partnership won’t be appropriate–if a small or charged amino acid ends up on a tRNA meant to carry a large hydrophobic amino acid, the combination will be ‘insufficiently sticky’, and EF-Tu won’t attach or will fall off too soon. If a large hydrophobic amino acid ends up on a tRNA ‘tuned’ for a small one, then the combination is ‘too sticky’, and EF-Tu may not let go when the time comes. Broadly speaking, only the ‘goldilocks’ combinations will function efficiently, and another path to error is minimized.
tRNA structures all the same: CCA adding enzyme
Every amino acid is joined covalently to ‘its’ tRNA by a linkage between the -COOH group of the amino acid backbone and the 3′ end of the tRNA. Interestingly, the same 3-nucleotide ending is found on the end of every tRNA, it’s 5′-CCA-3′. Fascinatingly, this ending doesn’t start there–it’s added by a special enzyme that–you guessed it–serves all tRNAs. This is a rare (and extremely cool) example of an ENZYME that dabbles in specific nucleic acid sequences. As with the ribosome and EF-Tu, since this single machine must ‘service’ all the varied tRNAs, they’d better all feel alike to it, so it relies on general structural features.
Interesting information about how this step also contributes to product-checking (advanced)
tRNA structures all the same: Pol III transcripts (advanced)
This one’s a little weird, so bear with me… Live every other RNA in the cell, tRNAs are produced by the process of transcription. Like (almost) everything else out there in the division between prokaryotes and eukaryotes, things are more complex in eukaryotes. Whereas in E. coli (prokaryote), one RNA polymerase makes everything, in eukaryotes (you), separate RNA Polymerases make ribosomal RNA (pol I), messenger RNAs (Pol II) and tRNAs and some other small RNAs (polIII). The twist relevant to the current story is that Pol III promoters (that part of the DNA sequence that ‘attracts’ a polymerase & tells it where to start) are internal to the coding sequence–i.e. the region of the gene that is transcribed into RNA must also (when in DNA form) specify an attractive ‘landing pad’ for RNA Polymerase III. Of course, those same nucleotides much be functional as part of the tRNA structure… and since there’s one kind of RNA Pol III, every tRNA gene must be the same in that each must have sequences that ‘feel good’ to it… and thus every tRNA structure must bear those marks.
tRNA structures all the same: tRNA splicing
We tend to make a much bigger deal about mRNA splicing, but it happens to tRNAs to. As with the splicing machinery itself, as with the ribosome and DNA and RNA polymerases, a single machine handles all comers. In this case, its underlying mechanism aligns nicely with our story here: it’s recognition of substrates is largely based on STRUCTURE–if something matches up as being ‘largely a tRNA’ but with a bunch of (RNA) stuff hanging off the anticodon end, then the extra is cut out and the remnants glued back together.
tRNAs are each unique
Now that they’re done being interchangeable, let’s look at how each tRNA must stand apart from the others
Each tRNA must have a unique anticodon
This one’s sort of the ‘duh’ piece; this is the core job of a tRNA–to ‘recognize’ (through specific basepairing) a partner codon in the mRNA, thereby delivering it’s cargo of a (specific) amino acid to the growing polypeptide. Of course, this is biology, so it’s a little cooler than that–because the one position of the anticodon (the 5′ position on the anticodon, which basepairs with the 3′ position of the codon or “3rd nucleotide” in it) is allowed to ‘wiggle about’ in order to find a fit, some of the tRNAs can form decent basepairing interactions with several nucleotide partners, allowing them to recognize a few codons. This utterly cool, totally amazing phenomenon was predicted by Francis Crick based on… thinking and doodling and patterns in the codon table. I’m sure a blog post will appear on this topic eventually :-). By the way, since he was British, he called it ‘wobble pairing’ rather than ‘wiggle pairing’.
tRNA structure each unique: tRNA synthetases
Besides specific recognition of codons by anticodons, the other essential piece to this whole ‘translation’ process is that somehow the ‘right’ amino acid must end up only with its ‘matching’ tRNA–and no other. How do the ‘tRNA synthetases’–who do the adding–achieve this? Well, since all tRNAs have essentially the same shape, there must be information ‘stamped onto’ that shape. And that’s exactly the story–while preserving the basepairing and length/loop constraints of the ‘general tRNA form’, different tRNAs have different sequences. Those of you paying attention are no doubt thinking “Well, the anticodon is the obvious place for these ‘synthetases’ to look to make their decisions!”. You’re half right–it’s the obvious place to look… and maybe half right again–about half of all tRNA synthetases ‘look’ (rather, contact) there. But not all. There rest ignore the anticodon altogether. The most straightforward and simplest example is that of the synthetase for tRNA-alanine in E. coli. This machine requires only one thing–a G-U basepair (yup, this is a ‘wobble pairing’) in the ‘acceptor stem’ (the double helix that lies ‘below’ the amino acid attachment site in the figure above; purple). If a tRNA HAS a G-U pair at this position, the tRNA synthetase that adds alanine will do so. If you change the G-U pair in a tRNA ‘for’ alanine, it will be ignore.
This brings up another interesting insight about the genetic code–there must be a ‘hands off my sequence’ policy for it to work. Consider: not only must the tRNA for alanine ‘display’ the appropriate G-U pair, but all others must encode some other pair at this position. So if anybody ever asks you ‘who in the cell knows the genetic code’, realize it’s a trick question–coding arises through the ‘cooperative’ efforts of a bunch of players, and if anybody messes up, the whole house of cards comes down.
I’ve doubtless missed some things, but this list alone is pretty amazing!