 Everyone teaches something about basepairing in Introductory Biology, but sometimes it’s just in passing, with no greater emphasis than the diversity of sugar variants receives. But basepairing is the wellspring of life itself and lies at the heart of each of the major informational transactions (DNA replication, transcription, translation) as well as a diversity of more recently-discovered processes in regulation. More and more, we’re also seeing it as the great keystone of new therapies. What are the core concepts and how can we teach basepairing in order to showcase all of these aspects?
Everyone teaches something about basepairing in Introductory Biology, but sometimes it’s just in passing, with no greater emphasis than the diversity of sugar variants receives. But basepairing is the wellspring of life itself and lies at the heart of each of the major informational transactions (DNA replication, transcription, translation) as well as a diversity of more recently-discovered processes in regulation. More and more, we’re also seeing it as the great keystone of new therapies. What are the core concepts and how can we teach basepairing in order to showcase all of these aspects?
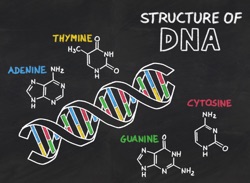
What it is: The core concept of basepairing is that the structures themselves can find and ‘recognize’ each other. No external machine is required to pair them up; our memorization (“A goes with T”) misses the point altogether. It’s critical that students understand that the shapes and ‘feels’ of complementary pairs ‘know’ one another, and that ‘erroneous’ pairs (Ade-Cyt; Gua-Thy) will reject their partners… all by themselves. The structure of DNA itself, on being witnessed, immediately illuminated DNA replication. More on basepairing and DNA in my blog post.
Replication: As Watson and Crick put it, “”It has not escaped our notice that the specific pairing we have postulated immediately suggest a possible copying mechanism for genetic material.” DNA polymerase is a machine for recognizing and cementing base PAIRS, not a hunter for ‘the partner of the one I have.” This gives rise to its noteworthy ability to synthesize DNA using nucleotides it has never ‘seen’ before (see this paper by Benner and colleagues). I think it’s critical that we communicate to students that thinking of the bases as letters is missing the entire boat. For more sophisticated students, it is interesting to cast the template base as part of the active site of DNA polymerase; specificity for each addition that polymerase arises from the existing strand. Some thoughts on the common “backbone part/variable part” of amino acids and nucleotides here.
Rant: I think there is a danger in getting WAY too caught up in the machinery of DNA replication, and missing the fact that all that stuff is just a ‘big helper.’ Life must have arisen in a nucleotide-like world with proteins as increasingly important adjuncts. If the list of all the (admittedly fascinating and currently vital) assistants displaces the roles of the bases themselves, I think we’ve lead students astray factually, and missed a trick both in terms of helping them understand the origins of life and in terms of understanding the essential simplicity of biological mechanisms.
Transcription: In many ways, there is no essential difference between replication and transcription mechanistically. Most of the keys that students are made to memorize are misleading (RNA can be as double stranded as DNA if it either folds to have secondary structure or somebody bothers to make the second strand) or are evolutionary embellishments (both the deoxy vs. oxy ribose difference and the use of thymine rather than uracil are advancements that allowed DNA to replace RNA as the long-term storage material in big genomes, but neither of these change the essence of pairing or information storage/transfer). Unfortunately, the purpose of transcription and replication are vastly different in today’s organisms, creating a minefield for new learners.
Adapters in translation (tRNA): Basepairing again rears its lovely head during translation. There’s a lengthy discussion in the link there, but the essence is familiar: the ‘information’ (instructions for making protein) in a messenger RNA must somehow come to ‘mean’ proteins. As Francis Crick intuited from first principles, there’s going to have to be a molecule with basepairing capabilities which has come to be linked to the specific amino acid to which its critical base sequence must correspond (yeah, he predicted synthetases too).
It’s worth pointing out that selection of reading frame involves a special case of tRNA:mRNA pairing. The match between an AUG codon and a methionine tRNA is the primary event in selecting where translation will start. And given that translation proceeds mechanically in groups of three from that point, that’s a pretty important decision!
Translation initiation (prokaryotes): Let’s not forget that tRNA-mRNA matching is not the only event in translation initiation. While eukaryotes have gone a different route (the 5′ cap and scanning), prokaryotes utilize basepairing between the Shine-Dalgarno sequence in the ribosome
Structure, structure everywhere: We don’t make a big deal of basepairing in RNA structure because it’s so… boring. But that doesn’t render it any less essential. tRNA and rRNA would be as useless as an unfolded protein enzyme if basepairing did not bring about their functional shapes (which in the case of rRNA, includes the enzyme peptidyl transferase).
structure of a teaching segment At some point, I’ll probably develop a general way of presenting these ‘modules’, but until then, I’ll just point out that I think they consist of three or four informational components (as distinct from delivery components). The stuff above is conceptual/underlying theory followed by broad thematic examples; next will be ‘real world examples.’ Unfortunately, I seem to be encountering two views on how folks understand things best. Personally, I’m “give me the concepts so I can hang my facts in an orderly way upon them” guy. My colleague Emily Dykstra is “Tell me something concrete, then I can incorporate your nebulous concepts around them.” I’ve met students matching each description, so I’m not sure there is a right-and-wrong here. At any rate, below are some “how cool is that?” instantiations where students can see how these principles are put into play making human life better, and perhaps providing them with gainful research employment or therapeutic tools in the future.
siRNA (small interfering RNA): A relatively recent discovery (by us, of course–organisms have known about this one for a long time 🙂 ). The chemical fact of basepairing means that any single strand of DNA or RNA is a ‘hunter-seeker’ capable of recognizing its complement and distinguishing it from all others, no matter how large the genome (given a large enough seeking strand). siRNA mechanisms include destruction of mRNA through double-helix formation and interference with translation by masking critical signals in mRNA. In an Honors class, I had students read “Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation.” (PNAS 109 #30 11975-80). Accompanied by a full lecture, a vocabulary accompaniment, and a guided-reading tutorial, it may’ve gone OK. As with everything I do, I’d double the investment and class time were I to try it again. If interested in my materials, drop me an e-mail.
CRISPR/Cas: Another basepairing-driven system, taken from basic study in biology but capable of being converted into a gene-editing tool (with a lot of commercial interest in doing so). As of this writing, I’m going to cop out and link to the relevant Wikipedia article and several articles written at a level that might be appropriate for Intro Bio if accompanied by guided lecture and/or worksheets:
New Scientist: Trying to edit out HIV
Technology Review: ‘Genome surgery‘
Kurzweil: ‘Replacing defective genes‘
NPR: A way to fix defective genes
Bits and Bobs
mutation: The fact that basepairing is ‘just’ a mechanical interaction between chemicals, mediated by precise positioning of H-bond donors and acceptors leaves it vulnerable to chemical processes. As long as genetic instructions are held in the form of carbon-based molecules, they’ll be subject to change anytime they’re in water. Amino groups have a bad habit of being replaced by oxygen, giving rise to adenine => hypoxanthine and cytosine => uracil conversions. These are deadly, as hypoxanthine partners perfectly with… cytosine, and uracil, of course, does so with adenine. That’s horrific news for maintenance of the ‘instructions’, in that the long-term result (if these errors aren’t caught) is a change from AT pair to GC or CG conversion to TA. Students can explore these changes using BasePairer.
origins: As alluded above, the fact and nature of basepairing are intimately associated with the origins of life… and the ability of life to originate. Whatever the detailed path of our origins was, there must have been a non-magical transition from something that we would identify as ‘chemical’ to entities capable of generating copies of themselves. The fact that the four bases are ‘elemental’ units encapsulating the copy-ability should not escape our notice… or that of our students!