Teaching biology at the cell/molecular level is challenging; as the Bob Seger song puts it ‘Deadlines and commitments/what to leave in… what to leave out.” How to decide? The glib answer is “identify the key concepts.” A definition of these might be “what are the aspects and ideas without which the process would not make sense, and which, if changed, would rock the cell’s world.” So… what are the Big Concepts of translation? Starting with negation, these cannot be the names or even the structures of the twenty amino acids (believed to be a combination of what was on hand in abundance ‘in the beginning’ and ‘discoveries’ that were biochemically accessible) nor their codon assignments, which, may reflect some physical correspondences, but seem to largely be a ‘frozen accident’ that we should not expect to find elsewhere. Rant: it cannot be the ability to ‘translate’ a message in a codons-to-amino-acids look-up table, as this has nothing whatever to do with the biological process! And the core cannot be a laundry list of actors and factors.
The big concepts in translation…
have to do with the processes by which living systems to convert from a language involving self-partnering chemicals–the bases— ideal for copying and dispensing ‘instructions’, to one that constitutes a wondrous diversity of building blocks–the amino acids— from which virtually any cellular machine can be made..
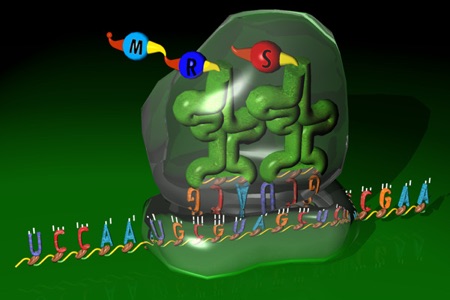
An attempt to represent a conceptual overview of translation is thinkBio’s Info Flow (Protein)
So herein lies the rub: we have two fantastic collections for their roles, one ‘informational’ and one action-based (pieces for building machines). Somehow, we have to get the instructions to instruct the assembly of the actors. Translation (going from ‘nucleotide language’ to ‘amino acid language’) is perhaps one of the most astounding and hardest to view as having evolved. Can we identify the root concepts in translation ? My version:
- The nature of the pieces #1 (bases): ‘Complementary matchers’ [the bases] that are intrinsically, structurally capable of being copied. No copier required. And if available in duplex format, reproduction is innate in the structure. Wow.
- The nature of the pieces #2 (amino acids): ‘Building blocks’ [the amino acids], a small number of which constitute a sufficient set of blocks to build… anything. No, really—anything! While I say above that the amino acid structures themselves aren’t a key concept, the diversity represented among them is. To name names, we have a tiny corner-turner (glycine), a fixed angler (proline) a smorgasbord of hydrophobic of various size and flexibility (Ala, Val, Ile, Leu, Met, Phe), some big rings of various properties (Phe, Tyr, Trp, His), a covalent link-maker (Cys), a pH-sensor (His), positive (Lys, Arg) and negative (Glu, Asp) specialists, as well as dedicated H-bonders (Gln, Asn). I think it’s fair to say that none of these individuals is essential in detail; if it’s functionality were achieved through a slightly different structure, all would be well. It’s interesting to point out shortcomings in passing: Cys is the only decent redox player, and nobody has a good metal for a sidechain (think heme in hemoglobin, or the irons and coppers of the electron transport chain).
- A way to invest sequences made of PIECES(#1) with ‘meaning’ in terms of PIECES(#2).
In other words, how do we take ‘meaningless’ but copy-able sequences of nucleic acid and ‘interpret’ them into sequences of amino acids? Initial proposals were that adjacent nucleotides of DNA formed ‘pockets’ for amino acid sidechains; Crick later predicted the existence ‘adaptor molecules’, with key like nucleic acid features, but with amino acids attached. Yes. PREDICTED tRNA. Oh, and in the same paper (actually, an informal letter to colleagues) predicted that there would be specific enzymes, each dedicated to a single amino acid–the tRNA synthetases. Yowzer.
- Ability of strings of PIECES(#2) [amino acids] to spontaneously take on forms capable of performing (infinitely) diverse functions (protein folding and from there, structure=function). Exactly where this concept goes is arguable, but if the linear strings of amino acids did not self-assemble into 3D functioning machines, the game would never get off the ground. Conceptually, all the machinery needs to do is ‘read out’ a nucleotide sequence as an amino acid sequence and ‘let go’, and the properties of the building blocks (amino acid sides chains as well as repeating backbone elements) will spontaneously give rise to folding.
If these are the broad concepts, what then is needed to understand the process? I think there is a broad principle of ‘mechanical specificity’–a series of machines each with a single lock-and-key type of role to play, and the a step back to try to see how the whole is so much greater than the sum of the parts. A much-appreciated lab exercise we do is based on a translation ‘play.’ Based extremely loosely on the delightful Stanford production from 1971, we drop the artistic elements and try to capture the essence of the players through magnet-studded blocks (hand manufactured by Kevin Baker and his home woodworking set-up; I could probably get his info!). I’ll integrate the explanation in the following ‘actor list’
- mRNA. Kinda obvious; to convert from language #1 to language #2, there must be a text to convert. Key features: AUG sequence (+ prok/euk specific features that help direct; prok = Shine-Dalgarno; euk = 5′ cap), codons, STOP codon
- tRNAs. (One per codon) minus (stop codons). Key features: anticodon, a region employing the base-matching aspect of the nucleotide ‘language’; recognition elements for synthetases to UNIQUELY find its target and not others [while it would be ‘obvious’ for the synthetases to ‘look at’ the anticodon, only about half do];
- synthetases (One per class of tRNA = one per amino acid). These sometimes get short shrift, but if anybody ‘knows’ how to translate, it’s these molecules. I often compare them to two-handed machines–one hand is shaped to grasp a specific amino acid; the second recognize structural elements common to a family of tRNAs that recognize codons ‘calling for’ that amino acid. Key features: specific recognition of an amino acid; specific recognition of tRNA family (some interesting notes: 1) as alluded above, some ignore the anticodon altogether! 2) the recognition ‘signature’ for the alanine synthetase is a single GC basepair in the acceptor stem [the ‘upper’ helix where the amino acid is attached] 3) for the code to ‘work’, not only must each synthetase have a ‘recognition signature’ that it recognizes, but all incorrect tRNAs must NOT contain that signature [this is the conceptual equivalent of “not only must A pair with T; A must not pair with C”], enzymatic capability, in that reactions are ‘uphill’, so ATP is used to ‘activate’ the amino acid to drive the amino acid-tRNA joining reaction, proofreading capability: this is a sophisticated idea, but fun to discuss: since the amino acid valine is essentially a perfect subset of the amino acid isoleucine, no pocket can be built that will recognize isoleucine that will not also recognize valine as a side-effect. By having a second pocket linked to hydrolytic activity that recognizes valine and excludes isoleucine, errors are quickly removed
- ribosomal subunits: As discussed in a previous post, this machine must be able to join any of the 20 amino acids to any other, so relies upon common elements of every tRNA. Key features: assembly using sequence signals in the mRNA (AUG and either Shine-Dalgarno sequence or 5′ cap); enzymatic capability to do covalent joining, ‘walking‘ capability to ‘stride’ down the mRNA in ‘steps’ of three (this is actually a combination activity with the Elongation Factors)
- termination factors: Molecular machines are dumb (or rather, mechanical). If nothing ‘tells’ the ribosome that it is finished, it will sit there until the End Days (or until Brownian motion wrests its two halves apart). So ‘Stop’ codons must actively mean stop. This role is carried out by termination factors, which are proteins that show astounding 3D mimicry of tRNAs (for the obvious reason that they must fit into the exact same pocket). Key features: can ‘read‘ stop triplets; can drive the disassembly of the ribosome
What makes all this fun to teach is that the machines themselves are each relatively simple (barring the ribosome, but it can be treated as a black box without damaging a conceptual approach, in my opinion). But by each machine doing a ‘simple’ recognition task, the net effect is… translation! I think we have an opportunity here to give students a deep and meaningful understanding of a process at the heart of all living things. But to do it, we have to focus on the key concepts in translation–what roles are necessary and how these roles are carried out by knee-jerk machines following simple, chemically-driven recognition rules…