Introductory biology can be overwhelming; what we consider to be ‘essential foundation’ could take up an entire undergraduate education. This issue is compounded when we treat the body of knowledge as a collection of names and facts (or when students achieve this view despite our efforts). By viewing the cell as a functioning machine, divided into compartments filled tasks performed by mindless but efficient mechanisms, a course can focus on understanding. A natural framework for student study and learning arises: the mechanics of the cell
Introductory biology can be overwhelming; what we consider to be ‘essential foundation’ could take up an entire undergraduate education. This issue is compounded when we treat the body of knowledge as a collection of names and facts (or when students achieve this view despite our efforts). By viewing the cell as a functioning machine, divided into compartments filled tasks performed by mindless but efficient mechanisms, a course can focus on understanding. A natural framework for student study and learning arises: the mechanics of the cell
While Scott Freeman’s ‘Biological Science’ has been my choice as textbook for my own Intro Bio course, some of its interesting decisions seem off base to me. The pre-printed highlighting of the text is itself a philosophy of learning. In looking at it, my summary would be “Learn the words and definitions; the sense will take care of itself.” I believe this to be the opposite of the case, and I think current thinking about how people learn backs this up.
Instead of approaching Introductory Biology as a body of information, I think good course design would have us dividing and conquering the mechanics of the cell into functional, purpose-fulfilling units. This view is one that students are already familiar with from their own lives, and thus parallels their world. It also demystifies many of the events and processes. As laid out in previous posts, I think most of life’s molecules are comprehensible to students and can be better approached and better retained if we talk about how things work and why they work that way (see previous posts on hemoglobin, opsins, timers, this exercise on tubulin…).
Meiosis: a sensible way to divide
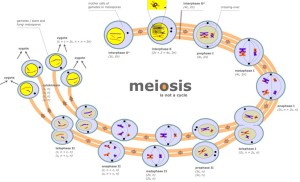
A machine-based view of the cell carries with it a framework-for-understanding. For example, in looking at meiosis, rather than having students memorize the steps, their characteristics, and how to recognize them in pictures, start from a viewpoint of “what are the cell’s goals here? What must get done?” If two gametes are going to unite to create a new individual of the same species, then the genetic complement of that individual must be equivalent to that of the parents. Lesson: each parent must contribute half of their holdings such that the offspring will have one full complement. Equally important, this cannot be just any half, but must be a half that will precisely complement the half contributed by the other parent. There’s only one way to achieve this effectively: each parent contains two ‘of everything’, and should create particles each holding precisely one ‘of everything.’. One of the best analogies I have heard for this process comes originally from Kim Nasmyth, and has to do with pairing socks: briefly, if I have a huge collection of items that I ‘know’ all exist as pairs, but they are jumbled (and for some reason, you and I each demand one of each pair), the best way to proceed is to find sock PAIRS and distribute one to each of us–a pretty fair summary of homolog pairing in meiosis! Similarly, once we replicate DNA in meiosis, we’re doomed to two divisions to achieve our goal of one-of-everything. Once we set a goal of achieving diversity in gametes, we require randomization events, and indeed find two–the fact that the orientation of each pair of homologs is random (the pole ‘faced’ by maternal-paternal homologs of chromosome 1 is independent of the orientation of the chromosome 2 homologs), and active exchange of genetic information to create ‘patchwork’ chromosomes. This exchange (homologous recombination) must in turn preserve the ‘exactly half’ plan, so the exchange will be between matched portions of homologs, not random swapping.
Mechanics of the cell: it’s how things work
Starting with a grand view of the (eukaryotic) cell as a compartmentalized whole gives students a familiar framework. I point out the advantages of membrane-bound organelles simply by asking them what they would think of an apartment where the boundaries (walls) of their bathroom, sleeping quarters, and kitchen were removed, and functions of each were performed in random locations. This analogy works especially well given that many students live with suite mates :-). This provides a SENSE to things, as opposed to making students memorize names, appearances, and events-occurring-within for each organelle. Other comparisons are also useful; the lysosome is the mechanical cell’s close equivalent to the stomach, all the way down to its low pH and harboring of digestive enzymes.
We should begin by tasking ourselves with always starting from a viewpoint of “how is this functionality important?” How is it achieved? What principles of (biological) machine design does it incorporate (can I extract a broader lesson? have I already covered this principle?)” I’m going to break out each of these, with examples, in order to illustrate how a ‘mechanics of the cell’ approach can be delivered.
How is this functionality important
Take membranes and their water-excluding property. As alluded above, I think rather than focusing on this property as part of the ‘list of characteristics of each biopolymer’, curriculum should be purpose driven. Once students recognize that segregation of functions (bathroom/kitchen/bedroom) is a valued part of their own lives, it’s a tiny step to look at the cell that way. The concept is easily expanded: the cell’s precious heritage of instructions is sequestered in the nucleus, hopefully protected from harm and located where it can be coddled by a host of repair and maintenance functionalities, and where ‘requests for reference information’ (gene regulation) can be sent, and the relevant (protein making) commands sent forth.
How is this mechanical property achieved
Two of my favorites here would be ‘information/instruction holding by DNA’ and enzymatic function of easing reaction likelihood. For DNA, as I outline in a previous post, I think we can lead students to the idea that the CRITICAL feature of an writing/coping/instructing process is high fidelity matching of units. This, in turn, calls for unambiguous partnering, whether same-with-same (as when we hand copy a letter) or same-with-other (as with G-C pairing or, I believe, with the ACTUAL inner workings of a Xerox machine).
To achieve unambiguous copying, we can extract several critical features the bases exemplify: rigid presentation of the ‘matching face’ (the purine/pyrimidine rings and their included double bonds give rise to pancake-flat, unyielding structures, thereby achieving reliable presentation of the H-bond donor/acceptor positions), binary matching types (partial (+) and (-) of H-bond donors and acceptors), and a very limited ‘character set’ (2 purines, 2 pyrimidines) that have perfect partners and horrible interactions with non-partners, allowing for clean, absolute “yes or no” matching.
For enzymes, instead of filling student heads with clouds of “lowering reaction barriers” and curves corresponding to nothing in their universes. Walking them through a straightforward chemical reaction and showing how the detailed structure of a protein (or rather, its 3-dimensionally optimized amino acid sidechains) gives a concrete look at how an enzyme enhances the doing of those things that need to be done for a given chemical reaction.
What principles arise from this example?
We need to be ruthless in selecting illustrative examples and tireless in deriving Big Principles from the selected examples. This post on translation attempts to do this. The pairing of DNA is involved in replication, transcription, ribosomal location of Shine-Dalgarno sequence (prokaryotes), tRNA-mRNA matching, identification of splice sites in pre-mRNA splicing, action of siRNAs in gene regulation, PCR, Southern blotting… The general regulatory principle of “To stop something, sit on a recognition site” occurs in the lac (and other) operons and eukaryotic gene regulation (see the design of the even-skipped regulatory regions), splicing, translational regulation, post-translational action driven by many protein-protein interactions… The regulatory principle of “to enhance a step, aid in the binding/presence of a positive effector” is common as well.
Since we don’t have the time even to teach all the classes of regulation or action well, much less the near-infinite number of instances, we best serve our students by indicating how carefully chosen examples illustrate principles that they should expect to see over and over… and over again in the mechanics of the cell.