Why combine protein function and modification? There’s a reason so many study sites have flashcard features, and it’s not just because they’re easy to program. I think textbooks and lecture can create the impression that introductory biology is nothing but a steady stream of equally-weighted, isolated terms and ‘things’. By finding and teaching relationships, we not only make big concepts easier to see and grasp, we increase the likelihood that they’ll stick and that students will feel a growing sense of power in their grasp of course material. This post will cover a unit that I think ties together a concrete example of enzyme function (ATPase), environmental effects and mechanism of pH (which is further explored in another thematic collection here), and protein control via phosphorylation (which, intriguingly, is achieved through a mechanism related to… ATPase chemistry!).
I’m always a bit taken aback to find a textbook that talks of amino acids and peptide bonds, includes proteins in the broad discussion of biopolymers, has an extensive discussion of nebulous things like ‘activation energies’ and the definition of a biological enzyme… and then sails away without looking at a single protein using its amino acids that are strung together by peptide bonds and spontaneously fold into a machine with an active site DOING protein things. This is worse than putting the cart before the horse; it’s skipping to the cart and ignoring the horse altogether. I think an orderly, comprehensible approach that starts with an ATP, includes ‘where the energy is’ in ATP, looks at how this ‘energy’ is locked up and released, and proceeds to consider how an enzyme can help gives students a concrete stepladder to climb up to the concepts we’re after.
I’ve dealt in a previous post with ATP and it’s packaged (i.e., potential) energy in the form of like charges locked in close proximity. I think that looking at accessing that energy can be very useful at the introductory biology level for a number of reasons:
- they’ll likely here of ATP a time or two before the course is over 🙂
- by understanding what holds ATP together, they can get an insight into how it falls apart
- once they understand why it doesn’t fall apart by itself (essentially, the barrier that must be overcome before the favorable reaction takes place) they are primed to see how an enzyme can help TAKE IT apart
- looking in detail at one such enzyme (using some beautiful graphics freely available here) ties together how enzymes work and makes concrete the idea that enzymes are just folded strings of proteins
- the underlying chemistry (SN2 attack) isn’t hard to wrap one’s head around and has a beautiful explanation here
- attacking phosphates is rather common, given that ATP is the ‘energy currency of the cell’, not to mention that phosphorylation is the most common form of post-translational modification. Consider these events, all driven by this same mechanism:
- breakdown of ATP to drive chemical reactions (yes–THAT reaction, that powers most of what the ‘energy currency’ is spent on
- polymerization of NTPs (3′ OH attack of triphosphate, with PPi as leaving group)
- phosphorylation of proteins on Ser, Thr, Tyr to regulate their activity
- if you teach G-protein coupled receptors (GPCRs), or the timer aspects of tubulin or EF-Tu, understanding the details of this reaction and how an enzyme enhances it, provides immediate insight into how one could ‘tune’ an enzyme to any rate desired… thus achieving a timer
Given an understanding of how enzymes work ‘buys’ us the right to talk about controlling them… and regulation has to be a Big Idea in introductory biology, however it may be presented. On the assumption that the course has covered how proteins fold prior to discussing them as enzymes or otherwise functional beasts, students should have a basic grasp that functions follows from form… and therefore to change form risks changing (or removing) function.
One could do the following pH-phosphorylation pair in whatever order suits the rest of the curriculum. As alluded above, I teach a pH module first that already would have introduced introduced the concept of protons as biological agents of protein-conformational change. Briefly, the highlights are that protons are like rain–if you go out where they are prevalent, you’ll get ‘wet’–protonated. Proteins are made of several different ‘fabrics’ (amino acids or more relevantly, ‘feels’), some of which are permanently ‘wet’ (lysine, arginine are almost always positively charged), some are ‘unwettable’ (constitutively negatively charged aspartic and glutamic acids) and one very special amino acid, histidine, wets and dries readily, depending on the concentration of protons in its environment–histidine is a ready-made pH sensor or pH-dependent switch. I tell students that pH is relevant to them because it’s a measure of a variable (environmental richness of H+) that influences a lot of biological processes and can vary in different environmental processes.
From here, I think we’re in a position to make a wonderful, sense-making linkage for the students about protein function and modification. pH is a ‘global effector’–in a given environment, all proteins receive the same amount of proton-bombardment, and one generally cannot change the environment of one protein in the mix to achieve differential effects. This works fine for situations like blood flow from high oxygen, higher-pH lungs to lower oxygen, lower pH tissues. And this is a major facet of how hemoglobin works. But what if you wanted to ‘reach out and touch’ a single (kind of) protein? What if you wanted your change to be quantitative rather than stochastic? What if you wanted your change to be semi-permanent (wonderful, brief review with students: what kind of bond would you make that has these characteristics?)? Answer to all these questions: you ‘glue’ on a charge via a covalent bond.
And that’s exactly what phosphorylation is: permanent attachment of a cluster of negative charges (phosphates) to a specifically-recognized protein motif (if you’re group is ready, you could of course go deeper into the idea that you could ‘define’ the specificity of a kinase as broadly or specifically as needed, depending on exactly what its target sequence was, and which proteins that sequence was ‘written on’ in a given environment).
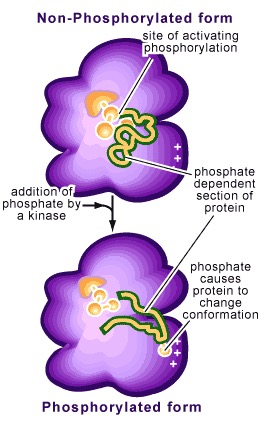
Below is a an image from a nice, basic-level review of phosphorylation, and is the source of the image below:
There’s a great ‘actual’ figure of this in Scott Freeman’s ‘Biological Science’ textbook. The key idea is straightforward: in principle, at least, the effect phosphorylation has on a given protein arises quite directly from a change in charge in one region without a compensatory change elsewhere, upsetting balances of power and preferred conformations.
A different-but-easily-understood example of protein function and modification arises in the ‘GTPase timers’, where the mere physical presence of the terminal (gamma) phosphate in GTP ‘gets in the way’ of some protein elements when it is present, but its removal creates space, allows protein elements to ‘move in’, and as above, drives conformational change. Here’s a wonderful, succinct video tutorial from Garland (released in association with Molecular Biology of the Cell textbook). Again, the trigger is ‘popping off’ a phosphate from a nucleotide triphosphate; again students can feel comfortable because they’ve been through the HOW and are equipped to think about the why.
Pingback: Dependencies and foundations: introductory biology design