 Too often, we seem to act as if first year biology students aren’t ‘smart enough’ to understand things and that it would be ‘easier’ if we just told them the names of things and the order in which they operate. This matches up with no modern model of learning and assumes some magical transformation happens during their sophomore year. Instead, I think we should start them on a diet rich in concepts and unifying themes and ideas. In this first post of what I hope will be a series, I’ll lay out what I think the Big, teachable ideas in the biological process of translation are.
Too often, we seem to act as if first year biology students aren’t ‘smart enough’ to understand things and that it would be ‘easier’ if we just told them the names of things and the order in which they operate. This matches up with no modern model of learning and assumes some magical transformation happens during their sophomore year. Instead, I think we should start them on a diet rich in concepts and unifying themes and ideas. In this first post of what I hope will be a series, I’ll lay out what I think the Big, teachable ideas in the biological process of translation are.
The name’s the thing
Students often confuse every aspect of transcription and translation, including just their titles. This is a shame, as they are both wonderfully descriptive. TranSCRIPTION is the process of re-writing (scribing), and corresponds to simply copying one string of nucleotides into another (I’m ignoring the U vs. T thing and what’s hanging off the 2′ position on the ribose because neither matters at this point). Translation in biology, on the other hand, corresponds precisely to its everyday meaning: going from the ‘language of precise matching’ or ‘language of copying’ (bases) to the ‘language of machine building’ (amino acids).
Send a thief to catch a thief
Amidst all the myriad details that students may encounter in introductory biology there are some recurrent themes; I hope to accumulate these in a later post. One that we see in translation is that the precision-matching feature of the ‘language of copying’ (the H-bond donor and acceptor ‘faces’ of bases) is used almost anytime one wants to recognize single-stranded nucleic acid. In translation, we see this…
- In the synthesis of the mRNA itself, arising directly from ribo-base matching to DNA template base
- In the recognition of the ‘start’ codon by tRNA during initiation
- (in E. coli and many other bacteria in the basepairing interaction between the RNA of the small ribosomal subunit and a sequence embedded in all mRNAs [Shine-Dalgarno sequence]
- In the interaction between each tRNA and the mRNA during translation itself
I believe that exceptions are potentially also informative. In translation, we see tRNA synthetases as PROTEIN factors recognizing tRNAs… but the synthetases have amino acid recognition as well as enzymatic chores, both perhaps best left to proteins. Termination factors recognize codons, implying a role for an RNA machine… but they, like synthetases, are charged with enzymatic functions leading to the disassembly of the ribosome.
The “nucleic acid to find specific nucleic acid” rule appears elsewhere as well. In splicing, the signature sequences at the 5′ end and branch point of the pre-mRNA are recognized via basepairing (with the U1 and U2 snRNAs, respectively). small interfering (si) RNAs that interact with mRNA to cause degradation or block translation are RNA sequences, as their name implies. Further, the relatively recently discovered mass of regulatory RNAs that influence gene control in eukaryotes are RNAs and achieve their ends through base matching.
This huge and ubiquitous collection of interactions means several things to me as a teacher. First, that the specific matching of bases must be what I design my “what are nucleic acids” module around, and second, that I must extract and identify this Big Idea in during introducing its appearances. Third, I personally argue that if I make the point well during translation, I think this means that I don’t necessarily have to cover mRNA splicing, which I see as a combination of three Big Ideas I cover elsewhere:
- Send an RNA to catch an RNA, as detailed above
- Everything that happens is a target for regulation (I make this point in almost every aspect of what I DO cover)
- Regulation is generally about two things: to enhance, you better enable the binding of an essential component (i.e. lac operon CAP/CRP + RNA Polymerase; i.e. eukaryotic transcription enhancers). To negatively affect a process, block a required element from getting where it needs to be (i.e. in lac operon, repressor overlapping RNA Poly binding site)
All for one; one for all
There are some interesting conundrums arising with translation in biology. The way I phrase some of these to students are “Why must all tRNAs be alike” vs. “Why must all tRNAs be distinct?” and “How can a single ribosome, which is conceptually one enzyme, do all 400 possible pairwise amino acid additions?” (i.e. gly-gly gly-ala, gly-arg…). I’ll deal with these sequentially; both potentially give students major insights into how the cell must accomplish its business, and make for a much readier recall of what can otherwise become simply additions to their list of memorizables.
tRNAs: different yet the same. Students light on the reason why tRNAs must all be different rather quickly. Each is charged with being the carrier of a specific amino acid (pun intended…), and thus must be distinctive in order to allow its cognate tRNA synthetase to know its tRNA. Functionally, a tRNA must be ‘defined’ by its anticodon since it is this region that must ‘read’ the codon(s) specifying its amino acid. So the ‘same’ part usually drops out quickly. The different bit is harder, but if you can get students thinking about the life & times of a tRNA, you can generally get them to realize that they all most fit into and function within the exact same ‘receiving pocket’ on the ribosome (indeed, they must function interchangeably while occupying A, P and E sites of the ribosome). Thus they must be similar enough the the ribosome can treat them all as though they were the same thing, whereas the mRNA and the tRNA synthetase must see them as distinctive.
I’ve actually dealt with the ‘how can one ribosome with a single active site join any of the 20 amino acids to any other?’ question before. Briefly, while we tend to focus on the sidechains of the amino acids, the backbone components are just as essential. The ribosome’s enzymatic functionality (reminder: RNA is peptidyl transferase!) actually only has to deal with joining amino groups to carboxyl groups (formally, to attacking carbonyl carbons while tRNAs serve as leaving groups). Since all amino acids have identical backbone components (overlooking a little irregularity with proline), the ribosome can blissfully do its thing and ignore the sidechains. I think by presenting in this way, we can turn a lesson about memorizing the names of the common elements of the backbone of an amino acid into an insight about solutions to the cell’s problems of building… and why they must be so.
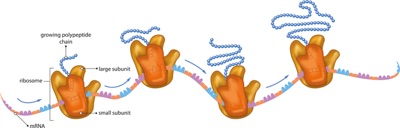
Adding who to whom
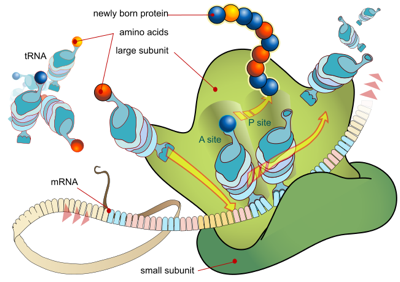
I often find students are confused about how actual progress is made in the growth of the amino acid chain. This is logical; as humans, we ‘add the new thing to the old thing’, be it in the building of brick walls or making sauces. So students are tempted to add the new amino acid to the existing chain. In the abstract, this is fine. In real life, it’s impossible and disastrous. I think this is a great opportunity to show students the power of concrete thinking–with chalk on a benchtop, on a whiteboard, on a piece of paper.
Attempts to add the new amino acid to the existing chain immediately give rise to several problems:
- If the new amino acid is added the way we do things with hands, the ribosome must ‘grab’ it off the incoming tRNA
- if it is added to the free end of the peptide chain, note that the free end is not held in a static position, and the number of places it could be increases rapidly as the length of the chain increases!
- If you are adding to the chain that is attached to the ‘old’ tRNA, and students recognize that the old tRNA is going to be exited, they’ll quickly realize that it exits with the chain, and the new tRNA is alone and empty!
A little examination and most students will realize of their own accord what the correct reaction/interaction must be: the new amino acid must steal the existing chain from the old one, which is a sterically identical reaction no matter how long the chain is, and does not introduce any challenges associated with who now has the chain!
If I don’t do it, nobody will
There are several fascinating aspects to the termination factors, but the first key idea to point out to students is their very existence. As humans, we tend to think that as soon as the protein is complete, of course the machinery would stop, and as soon as the ribosome is done making protein, of course it recycles in preparation for a new round. Neither of these has meaning to molecules. Even in the absence of a matching tRNA for a STOP codon, the ribosome has no sense of time–it would cheerfully sit over the stop codon through thousands, millions of tRNA ‘visits’. The only thing to bring a halt to the madness would be the wrecking ball of Brownian motion. If the cell needs a process to stop, there must be a stopper. In this case, it’s a termination factor. Similarly, if the ribosome needs to go from an assembled state to a disassembled state, there must be a disassembler… again, our friend the termination factor.
A fascinating idea is that students can be lead to propose on their own is the structure of the termination factor. If a termination factor is to ‘read’ a stop codon, if it is to enter the ribosome portal that normally houses a tRNA, then it must basically look like a tRNA. And boy does it (point of nomenclature: termination factor = release factor = RF3)! Actually, the best resemblance is to tRNA + EF-Tu, which is to be expected as a tRNA is generally chaperoned by EF-Tu when it arrives at the ribosome. Note the image includes EF-G, the factor that moves the ribosome down the mRNA after each addition, and which bears the family resemblance for the same reason as (tRNA + EF-Tu) and termination factors.
It is important to point out the remarkable overall structural similarity of the tRNA portion of (EF-Tu + tRNA) and the release factor and EF-G–the first is made of RNA, the latter two of protein. This similarity brings home some of the arguments we each make that an “RNA world” is plausible, in which RNA played both critical roles–informational molecule and functional molecule.
Pingback: Pull yourself together! Self-assembly in biology