 Understanding the cellular roles of DNA and protein is challenging because they are, in important and meaningful ways, not analogous to anything in the macroscopic world. Proteins are a fascinating study: while cars, watches and houses are built by specialized builders, a single master factory, the ribosome, strings together all cellular proteins. And in a richly meaningful sense, that string then assembles itself (folds) into its final form. And the machines of the cell are almost unimaginably diverse in their forms and functions, the latter of which derive once again from within: the amino acid building blocks and precisely where they come to rest in the final three dimensional structure.
Understanding the cellular roles of DNA and protein is challenging because they are, in important and meaningful ways, not analogous to anything in the macroscopic world. Proteins are a fascinating study: while cars, watches and houses are built by specialized builders, a single master factory, the ribosome, strings together all cellular proteins. And in a richly meaningful sense, that string then assembles itself (folds) into its final form. And the machines of the cell are almost unimaginably diverse in their forms and functions, the latter of which derive once again from within: the amino acid building blocks and precisely where they come to rest in the final three dimensional structure.
The central mystery of life is that its primary machines have neither builders in the sense that we think of them nor overseers to coordinate their actions. Molecules are blind and dumb, with their only meaningful interactions being ‘touch’ (inter- and intra molecular binding). Understanding biology at the cellular level, then, requires a deep understanding of how DNA codes ‘for’ anything and how proteins begin life as ‘beads on a string’ (a necessary consequence of protein creation through linear ‘read out’ of an mRNA string) but come to be the awe-inspiring machines by which the cell’s needs are met and its functions carried out.
If you link it, they will assemble
One of the central solutions to the problem is that proteins are self assembling (my post on self-assembly as a universal theme here). The importance of this concept cannot be underestimated, though I think it’s a challenge to get students to understand the need for this property, and thus the wonder of scope of its solution can be lost. In introducing the idea, a brief visit to ‘infinite recursion’ can be useful: if machine A had a special machine that assembled it (A’), then the assembler (A’) would be a machine requiring, in turn, its own mechanical assembler (A”); this last would be just another in the chain, requiring a builder of its own (A”’), and so on and so on ad infinitum. The only way that life could arise from non life is if the instructional molecule (nucleic acids, at least nowadays) had a structure that made it innately self-copying (basepairing partners ‘know’ one another through charge-charge complementarity). The only way translation could be ‘bootstrapped’ is if complex machines (proteins) can arise from ticker-tape readouts that somehow achieved function from nothing more than the sequence of joined elements (on this planet, amino acids).
Life would be wondrous enough if proteins were ‘merely’ art–strings of a finite number of blocks that, dropped into water, spontaneously assemble into compact, complex three-dimensional shapes with various common motifs (alpha helix, beta sheet)… and doing this “first time, every time.” (in quote marks because that’s an ad campaign of some sort from my youth). But there’s so much more. Proteins must not only ‘look good’, they must perform any and every action and task the cell needs to survive (barring the occasional but tremendously important ribozyme or RNA machine). Even proteins that are ‘merely’ structural generally are regulated in their assembly/disassembly (don’t ever tell anyone that tubulin is a ‘simple’ filament!) and charged with interacting with a variety of other components.
From this setting, I think the amino acid building blocks can be introduced to students as a collection not of twenty annoying stick figures for memorization, but components of a toolkit, part accidental-historical and part needful (it seems reasonable to conjecture that some specific roles will also be found in independently evolved systems in other universes, though the detailed structures may differ). In my own courses, I try to create (and call attention to!) interesting instances to point out the special features that some of the building blocks contribute to the whole.
Everything isn’t a nail: 20 players for many parts
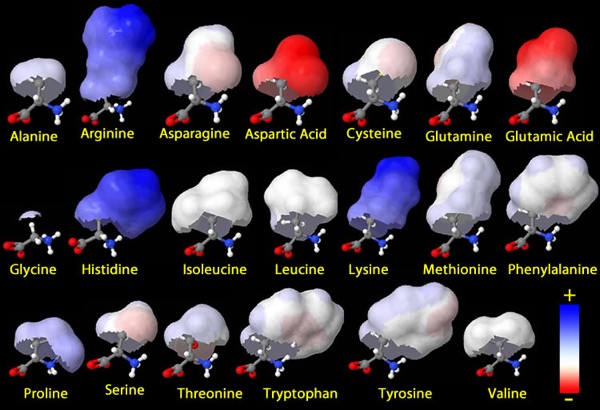
Of course, there are the familiar divisions students are often called upon to recognize–positive charges (Arg, Lys and sometimes His), negative charges (Asp, Glu), H-bonding specialists (Asn, Gln though others can play)… since these divisions are familiar, I’d like to emphasize several other aspects that I think bring the “wonderful toolkit” point of view better into focus and provide a good framework for lasting student understanding. So the following will be a tour of some amino acid building blocks who are called upon to step up and play starring roles because of the unique capabilities they bring to the team.
Histidine: pH sensor and metal binder
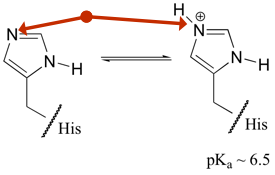
At first glance, histidine just looks like it was designed to have a hard-to-memorize structure. But watch out for that (shown-as-non-protonated) nitrogen!
http://chemwiki.ucdavis.edu/Organic_Chemistry/UMM_chemwiki_project/Acid%2F%2FBase_Reactions/The_Henderson-Hasselbalch_equation
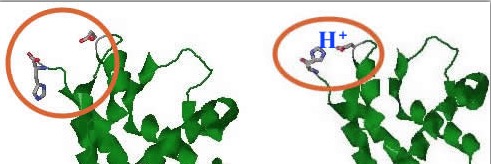
There are two keys to histidine’s ‘special powers’, both hinging on some complexities of the ring environment and the fact that two nitrogens are essentially symmetrically disposed within the ring. But enough about that–what are the consequences? Number one, it means that the presence/absence of that proton is determined by pH… Of course, the presence/absence of pretty much any hydrogen-on-a-nitrogen depends on pH, but the ‘balance point’ (=pKa) for this one is about 6.5–in other words, the point at which about 50% of histidines will HAVE the proton (and be positively charged). Without getting embroiled in issues of equilibrium and such, the Big Idea is that as pH heads lower, the odds of that histidine becoming a positively charged entity increase, and vice-versa. Who cares? It may not have escaped your notice that your own pH is in that range as well. In other words, histidine is perfectly placed to change its charge in response to environmental changes. This becomes particularly interesting when you realize that for a protein such as hemoglobin, the environment where it should behave one way (oxygen binder; lungs) has a different pH than a place where it is meant to have a markedly different behavior (release oxygen in tissues, where pH is lower). There are several interactions within histidine that are driven by presence/absence of protons, generating the ‘Bohr effect’. A histidine-glutamic acid interaction (and its equivalent non-interaction) from two states of hemoglobin is shown below (histidine on the left):
More whimsically (but making for a great classroom demonstration) is miraculin, a protein that sits on the tongue’s sweet taste receptors. Several histidines in the protein render it pH sensitive; at neutral pH, it just just sits there for a while. At low pH (such as occurs when biting into a lemon 🙂 ), it activates the sweet receptor, generating a wonderful lemonade without the sugar! thinkBio resources for an interesting paper on miraculin can be found in this table; the paper deals with identifying the residues that determine target specificity (mice don’t respond to miraculin), though a link in that paper goes to the research indicating that histidines at positions 30 and 60 mediate the pH-sensitive response.
But wait! There’s more to histidine. The lone pair electrons (where the proton joins at lower pH) are also just what some metal ions are looking for. One of the functionalities not found in the 20 amino acid building blocks is redox capability. Since metals are wonders in this department, the next best thing is to grab one. Thus we find heme or similar in hemoglobin, electron transport chains, chlorophyll, etc. And guess who gets the difficult job of playing nice with the metal? In the cases I’m familiar with, it’s our good friend histidine again. For a quick look, follow this link and examine alpha His87 => Tyr and beta His 92 => Gln.
In a tight spot: glycine
Glycine and its side chain are more amenable to analysis, since there simply isn’t a sidechain to be analyzed. However, this ‘feature’ is one readily grasped by students looking at the various twists and turns and overall tight packing of proteins. By being essentially a blank spot in the backbone, glycine nonetheless fills a valuable niche by making more structures possible than would be without it.
Round the bend: proline
Proline is unique among the amino acids in that formally speaking, it isn’t one–it’s an imino acid because the backbone nitrogen is attached to the terminal carbon of its sidechain. Obviously this constrains the ‘wiggliness’ of the sidechain as well as forcing the backbone into a fixed and reliable configuration. Avoiding discussions of entropy and such, it suffices to say that being so constrained renders proline uniquely good for some things and bad for others–such as being in alpha helices, something prolines studiously avoid.
Ties that bind: cysteine
Cysteine represents another fun use-case for a specialized amino acid. While it can be looked at as a ‘cousin of serine’ due to overall structural similarity and sulfur’s appearance just beneath oxygen in the periodic table, a more noteworthy feature is its willingness to form disulfide bonds when juxtaposed with other cysteines. Broadly speaking, this integrates well with general discussions in Introductory Biology, where almost everyone makes distinctions regarding the permanence and ‘strength’ of bonds as well as the vulnerability of proteins to denature under environmental stress. While all other interactions that drive folding are relatively weak non-covalent ones, well-placed cysteines provide an opportunity to ‘glue’ regions of a protein together, allowing them to better resist challenging environments.
An interesting appearance of this capability in experimental form is the ‘Scrambled ribonuclease’ experiment, where this feature of cysteines was used to ‘lock’ denatured ribonuclease in its unfolded state and then use this ‘proven dead’ form to show that the protein was capable of re-folding with no other factors present. Briefly, the protocol was
- Purify ribonuclease; demonstrate that it has enzymatic activity
- Reduce disulfide bonds (detach them) using beta-mercaptoethanol (common lab reagent that smell like skunk due to its own -SH moiety)
- Heat the protein and ‘snap chill’ it, forcing it to ‘fall into’ random, poorly folded structures
- Remove the reducing agent, thereby allowing the disulfide bonds to re-form–‘locking’ the protein in its poorly-folded state
- Demonstrate that the current form has lost essentially all enzymatic activity
- Take this purified, wholly inactive form, re-break the disulfide bonds, provide heat and slow cooling to favor renaturation
- Demonstrate that enzymatic activity has been restored with nothing in the bottle but ribonuclease itself–thus all the information for correct folding is within the protein!
While this will someday be covered in a Lectures => Great Papers => Scrambled ribonuclease link and hopefully a blog post, a little information and link to the paper can be found here.
I have taught this as part of an Honors adjunct to Introductory Biology; the paper is tough sledding because of the indirect methods required at the time, but I believe with sufficient discussion of methods, figures, vocabulary and overall guidance, success can be had.
Put a (charged) lid on it: serine, threonine, tyrosine
While it’s certainly reasonable to discuss these three in the context of H-bond donors and acceptors, I believe post-translational control via phosphorylation is far too important not to acknowledge this group in that light as well. As such, they represent “on/off” switches that can be woven into essentially any protein and depending on surrounding sequence/structure and specificity of kinase/phosphatase, made to respond to as specific or general a signal as desired. In a previous post, I discuss this as conceptually an extension of pH (see histidine above) in that pH provides general control of any affected protein and requires environmental differences, whereas phosphorylation affects only specified target(s) and can be done anywhere, anytime.
Of course, it should be noted that lysines get methylated, hydroxyproline is generated in collagen, etc.–as in all of biology, there are myriad variations on the theme…