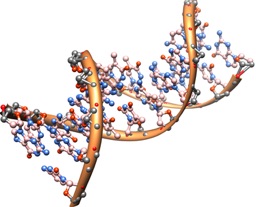
We use a several terms in IntroBio that all mean the same thing, but may mean little to students in the classroom–‘binding’, ‘specific interaction’, ‘interacts with’, ‘recognizes’. This is particularly dangerous with ‘recognizes’, because newcomers can envision a police line-up: targets appear and we visually choose one. Alternatively, we risk students imagining a somewhat mystical, or at least poorly-defined ‘way’ that molecules have that is beyond their ken. Nothing could be further from the truth; in some ways one of the easiest things to understand is why two molecules ‘stick to’ one another and overcome the force of Brownian motion, at least for a little while.
(Image source: http://imgur.com/PE07XGM)
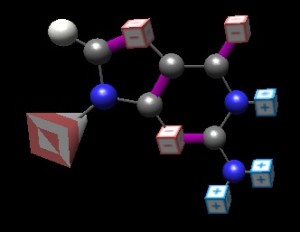
 Too often, biology is presented as gathering data and letting it lead us. While this is a valid and important component, there are also many glorious examples where the thinking came first and directed the experimentation. Many of the great experiments in biology came about this way–Meselson and Stahl (semi-conservative replication), Hershey-Chase (the Blender experiment, or ‘who is the genetic material)… and some that are more abstract–Pauling’s proposal of the alpha helix and beta sheet… and a number of Crick’s hypotheses. This post covers his prediction that efficiencies would be achieved in the tRNA pool through wobble pairing: relaxing orientation of the participants in a basepair to allow one base to partner with several others.
Too often, biology is presented as gathering data and letting it lead us. While this is a valid and important component, there are also many glorious examples where the thinking came first and directed the experimentation. Many of the great experiments in biology came about this way–Meselson and Stahl (semi-conservative replication), Hershey-Chase (the Blender experiment, or ‘who is the genetic material)… and some that are more abstract–Pauling’s proposal of the alpha helix and beta sheet… and a number of Crick’s hypotheses. This post covers his prediction that efficiencies would be achieved in the tRNA pool through wobble pairing: relaxing orientation of the participants in a basepair to allow one base to partner with several others.  Both studies and common sense indicate that common threads running through our teaching provide students with reinforcement of both thread components and the things they connect. This approach also highlights conveying principles rather than fact collecting as our learning objective. The details of a basic ATP hydrolysis reaction illustrate both key principles (how enzymes actually implement their abstract aspects [speed up reaction; lower reaction barrier], roles for specific amino acids and protein folding), value of understanding chemistry in thinking about biology) as well as providing students with a tool that they’ll see over and over… and over again: the core mechanism is found in nucleotide addition, kinase and phosphatase reactions, pre-mRNA splicing, the timing mechanisms of GTPases [tubulin, EF-Tu, GPCRs], and… oh yeah: virtually every ATP-driven or -coupled reaction in the cell!
Both studies and common sense indicate that common threads running through our teaching provide students with reinforcement of both thread components and the things they connect. This approach also highlights conveying principles rather than fact collecting as our learning objective. The details of a basic ATP hydrolysis reaction illustrate both key principles (how enzymes actually implement their abstract aspects [speed up reaction; lower reaction barrier], roles for specific amino acids and protein folding), value of understanding chemistry in thinking about biology) as well as providing students with a tool that they’ll see over and over… and over again: the core mechanism is found in nucleotide addition, kinase and phosphatase reactions, pre-mRNA splicing, the timing mechanisms of GTPases [tubulin, EF-Tu, GPCRs], and… oh yeah: virtually every ATP-driven or -coupled reaction in the cell!