A Great Experiment should have several qualities: it should address a Big Question of its time, it should ‘move the ball’ a good ways and be universally seen to do so, it should entail an approach that is insightful and hopefully clever, and should represent a careful foundation of what system and techniques should be employed–or invented. “Mutations affecting segment number and polarity in Drosophila” by Christiane Nüsslein-Volhard and Eric Wieschaus is just such a paper. FYI, the Lasker Award folks and the Nobel Prize selection committee thought so too. It represents a combination of a huge question (where does multicellular form come from given a single-celled start?), brilliant recognition of ideal experimental system (Drosophila combines ancient genetic tools with a ‘body plan’ that is scribed on the surface of the early embryo), invention (I believe) of Drosophila ‘replica plating’ and resulted in a series of mutants that by their existence and phenotypes left us with a clear idea of how the body plan of Drosophila is laid out through sequential gene-expression driven subdivision into smaller regions.
A Great Experiment should have several qualities: it should address a Big Question of its time, it should ‘move the ball’ a good ways and be universally seen to do so, it should entail an approach that is insightful and hopefully clever, and should represent a careful foundation of what system and techniques should be employed–or invented. “Mutations affecting segment number and polarity in Drosophila” by Christiane Nüsslein-Volhard and Eric Wieschaus is just such a paper. FYI, the Lasker Award folks and the Nobel Prize selection committee thought so too. It represents a combination of a huge question (where does multicellular form come from given a single-celled start?), brilliant recognition of ideal experimental system (Drosophila combines ancient genetic tools with a ‘body plan’ that is scribed on the surface of the early embryo), invention (I believe) of Drosophila ‘replica plating’ and resulted in a series of mutants that by their existence and phenotypes left us with a clear idea of how the body plan of Drosophila is laid out through sequential gene-expression driven subdivision into smaller regions.
In teaching students ‘authentic’ science, I think there are a number of things we must convey. My own view is that especially at the introductory level, ‘cutting edge’ papers in molecular biology are essentially inaccessible (massive jargon, intimate + extensive background required, case-specific details of methods, rarely universal questions). If that’s the case, then we’re going to be dealing with curated examples from the past. Translation: it can’t be about the conclusions; these are already in student textbooks and may well be part of the litany (the proton motive force powering mitochondrial ATP production, elimination of replication models other than semiconservative, demonstration that the codons are 3 nucleotides). So what must such experiments be about? I propose they should be exemplars of at least some of the following:
- What was the Big Question, and why was everyone (or many ones, at the very least) thinking about it?
- Why didn’t we already know the answer? What were barriers to solutions?
- What were the competing hypotheses?
- Why was the experimental system chosen? (*I think this often separates great experiments from mundane ones, and failures. And I think these decisions are often invisible to students)
- What is the path from the experimental RESULTS to the death of discarded MODELS? (my view is that students learn that experimentation is merely a confirmatory tool because we tend to teach result before question)
- Why did the entire field advance upon acknowledgement of the results? It can’t be a great experiment if it didn’t accomplish anything!
So, let’s examine the case of the Wieschaus-Nusslein-Volhard drosophila development Nobel experiments. Disclaimer: I’m neither a developmental biologist nor a Drosophila dude, so this is an awed-outsider-looking-in post.
The Big Question
This one’s easy–broadly stated, how does a single cell (zygote), by definition of only one ‘type’, give rise to the ordered, diverse body plan seen in so many astounding eukaryotes? We’d pretty much imagined the homunculus-in-the-sperm idea (progress!). Models involving diffusion and gradients had long gotten interesting play, with noteworthy successes such as limb development (for a nice recent (i.e., non-period-relevant) review, I like the look of this article).
References
“The Heidelberg Screen for Pattern Mutants of Drosophila: A Personal Account” Eric Wieschaus and Christiane Nüsslein-Volhard, Annual Review of Cell and Developmental Biology
Vol. 32:1-46 (Volume publication date October 2016)
***A wonderful review of the science, the data; has great images and accessible explanations
“Mutations affecting segment number and polarity in Drosophila” C. Nusslein-Volhard & E. Wieschaus, Nature 287 (30 Oct. 1980) 795-81
“Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster I. Zygotic loci on the second chromosome” C. Nusslein-Volhard, E. Wieschaus and H. Kluding, Roux’s Archives of Developmental Biology 193 (1984) 267-82
Best line from the third paper: “…after which the parents were discarded with the help of a vacuum cleaner.”
Answers before questions: Freeman fly-building tutorial
A lecture by Prof. Wieschaus describing the mutant hunt from the ground up… what really happened
Existing knowledge/barriers
I think the key insight to this work is that the authors established a path to the answers. The scope of that idea is important here. The foundation is that a system displaying the phenomenon is needed, so the yeasts and other beloved single-cell systems are right out. The facilities and generation times for mouse or zebrafish would be terribly limiting (and I’m not sure zebrafish was even a thing yet).
I’m going to cover this more in depth under the ‘Experimental system’, but the choice of Drosophila is really what makes this work. The authors started with the sensible premise that to learn about development genetically, they’d be looking at mutants that failed in the process. They were going to be dead. Their huge insight was that a developing embryo “wears its pattern on its sleeve” as it were–the pattern of denticles (hairs) and other ‘signature’ aspects of segment identity appear on the surface of the embryo. For a manual mutant-screening process, this is huge. No dissection, no fancy staining protocols, just look at the dead dudes and listen to what they’re telling you (I’m oversimplifying here; the Materials & Methods section is not actually ‘put under microscope; profit!’).
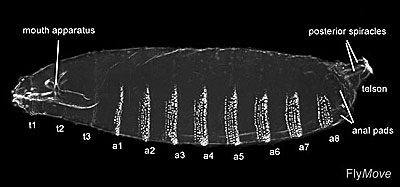
From http://flymove.uni-muenster.de/Glossary/GlossPics/Cuticula.jpg
Even from this point, though, more innovation was needed. On the assumption that their target was recessive lethals, they were going to need to be generating lines of mutants so that homozygous recessive lethals were there to be observed–and stayed there (alleles that cause death tend to disappear from populations :-)–more under ‘system’). And they wanted to look at the embryos that had died, and distinguish them from those dying of other causes. Overall, we’d better hope for one of the most established genetic systems on the planet.
The hope had to be that developmental mutants would be ‘interesting’–i.e. informative. You can imagine that every failure of pattern-formation would give rise to disaster, and hundreds of hours would’ve been spent generating different ways of making Drosophila stew. I’m pretty sure the study would not have been embarked upon had the authors not held out hope that they could do better.
Competing hypotheses
Being an outsider and lacking someone else’s insightful review, I’m not going to go further than to say that it appears to me that the frontier was largely open at this point. The authors were doing more ‘exploratory’ science, taking the approach of “Let’s force the organism to show us all the ways it can fail to create itself, then we’ll look upon its failures, classify them, and develop a rule-based system for what it’s doing.” (Hindsight indeed has its joys!)
As alluded above, there was a possibility that “it’s gradients all the way down”; evidence already indicated that some of the maternal contribution looked to have a gradient type nature. One could also consider several possibilities for how things worked; I’ll throw out a couple:
- Each segment’s identity was the expression of a single gene or set of genes specific to it
- There was sequential ‘addressing’ from front-to-back or vice-versa
- segment addressing is somehow simultaneous (this would accord with gradient models)
- The actual case, where a time-driven ‘cascade’ of (often binary) divisions (beginning with chunks not formally related to the final pattern) segment identity is ‘assembled’ as each segment represents a unique set of gene product presences
Experimental system
Drosophila is not my favorite organism; too many have flown up my nose for that. However, it’s hard to argue that the fruit fly is not the perfect marriage of historical power and question-appropriateness for the authors’ quest. Let me break down these airy claims:
Heart on my sleeve: the segmental body plan and denticle pattern. From start, the authors were going to have to look at literally thousands of potential mutants. Many of these would not even be mutant; many others would not be helpful to their cause. The ability to (relatively) rapidly recognize desirable candidates is the name of the game here. The fact that the state of the Drosophila embryo can be assessed by viewing it (once the egg has been stripped away chemically, a process that sounds to be at least approachable en masse). So an expert could tell by visual assessment how the developmental process had worked out for a given embryo (again, kudos to them–I look at their published embryo pictures and see a mess, though I can imagine the denticle ‘signatures’ provide the needed specificity).
Goldilocks of death: the potent Drosophila genetic system was also used to handle several mundane aspects of their mutant hunt quite elegantly. Briefly, they knew the mutants they wanted would die when homozygous. That means that stocks containing their mutations would steadily lose the very mutations they sought unless something were done. Further, these being eukaryotes and all, recombination would could move the mutations to homologous chromosomes ‘un-linking’ them from genetic markers used to follow them. Both issues were elegantly solved by taking thoughtful advantage of multiply (genetically) marked Drosophila ‘balancer’ chromosomes.
Balancer chromosomes are chromosomes that have undergone many massive rearrangements, essentially preventing fruitful recombination with homologs. Further, the wizards of Drosophila have produced ones that have both dominant-interesting phenotypes (i.e. give rise to ‘curly’ wings), recessive-notable ones (white instead of red eyes), and (drumroll please), recessive lethal alleles. Let’s play out what this means for maintaining the mutant lines presumably bearing a lethal developmental mutation:
- Because the balancer chromosome is intensely rearranged, all genetic markers on the balancer stay ‘there’, and the recessive lethal developmental mutation is ‘locked’ onto its own chromosome
- Because the balancer chromosome bears a recessive lethal mutation, every generation the homozygotes of EACH chrosome alternative (balancer/balancer or dev mutant/dev mutant) die out; you essentially have a true-breeding line of heterozygotes. Try that, essentially any other genetic system!
- Because the balancer chromosome bears a DOMINANT phenotype-conferring allele, visual inspection of each line can confirm the continued present of the balancer chromosome
- Symmetric to the case above, if the line is heterozygous for a phenotype-generating allele with the dominant on the ‘mutated/developmental mutant’ line, the presence of the mutagenized chromosome can also be visually assessed
Oh, by the way–a separate system and screen had to be performed for each of Drosophila’s big chromosomes (X, 2 and 3); they skipped 4 because it’s puny enough to be not worth the trouble! FYI, I’m not sure if the primary results from X and 3 were published; the references at the top of this post are for the big picture (Nature article) and the chromosome 2 hunt.
Path: Results to Conclusions
To me, this is where this work really shines. Massive amounts of labor go in; innovation, careful selection of system, massive amounts of labor (yes I already said it, but it’s worth repeating!). What comes out of all this?
We know how it works! Drosophila development proceeds as stepwise divisions of the embryo in a cascade of ever-finer divisions, most of which depend on positional information generated by previous steps. That sentence likely isn’t worth much, so I’ll break things down a bit.
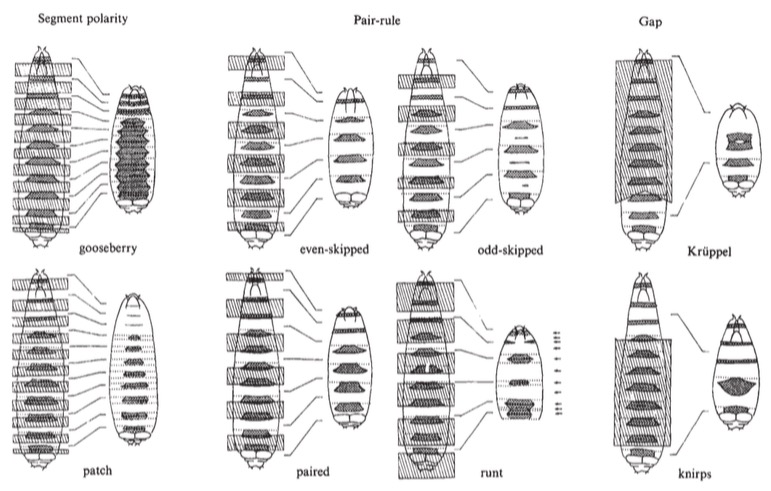
First and foremost, a couple sentences on how geneticists think. The basic philosophy is “staring at a working machine may not tell me anything. Let me break one piece and see how the machine fails; I will then become aware of one of the things that the machine had been doing, I will have a handle on the thing that did it [it’s now broken, which is an entryway], and I will have an insight into what happens in the absence of the broken thing”.
In the image above, for each pair of images, the leftmost is a complete embryo with the bits that will be removed in the absence of gene function of a given mutant; the right image in the pair shows the resulting embryo. Thus knirps, lower right, is missing a huge chunk from about the 1/3 mark to the 5/6 mark, leaving a student embryo with few bands of hairs (denticle bands) still present.
As alluded in the ‘Competing models’ segment, several things are apparent. No gene has a one-to-one correspondence to an eventual body segment. The function of some genes is required over large contiguous swaths of the body; others act repeatedly, often in corresponding regions of each segment (gooseberry, patched) or every other segment (even- and odd-skipped). The emergent picture is one in which the gene products must be functionally acting as ‘markers’ of territories or ‘addresses’.
The mutations themselves suggest a hierarchy, with some acting before and providing positional information to others. This has been borne out in subsequent work. Some of the most complete has been on the eve (even-skipped) gene, where it has been shown that each segment where even-skipped is expressed (and thus each segment that fails to properly appear when even-skipped is mutated) uses a different upstream controlling DNA element, allowing it to ‘construct’ its decision based on the different positional inputs (presence/absence of other gene products) where a given stripe must appear.
Analysis of even-skipped expression (wonderfully laid out in the free [as long as you only want a chunk during any given visit] on-line textbook Alberts et al.]
“Transcriptional regulation of a pair-rule stripe in Drosophila” Small S., Kraut R., Hoey T., Warrior R., Levine M. Genes and Development 1991 5 (5): 827-39
Everybody moves forward: Drosophila development ‘solved’
The image above is a delightful example of how to present data in a way that illuminates models and causes thinking. It’s utterly clear that a major aspect of body plan mapping in Drosophila is based on defining specific regions. Further, there is a clear suggestion of hierarchy even before experiments involving time-of-action and epistasis (which of two mutant phenotypes ‘shines through’ when both mutations are present) experiments were done. Anyone looking at the figure would infer that the definition of the larger chunks precedes, and is likely the basis of the smaller subdivisions. And this has been borne out.
One of the joys of a ‘saturating mutant hunt’ (i.e. one in which the hunters attempt to get so many mutants that they recover all the mutants that could possibly be gotten) is that it allows us to think about the entire set. The authors make statistical arguments (based on the number of duplicate instances of recovery of some genes) that while they did not recover the complete set of genes matching their search criteria, they identified the majority. In other words, the above is not so much a view of ‘some of the things that go on in ordering the Drosophila embryo (as constrained by the authors’ methods, of course) but rather most of the events involved in this aspect of Drosophila development. While some of the mutants recovered had been known before, a (semi) complete set allowed us to understand their place in the grand scheme and to think about whether they were typical of a class of genes.
Of course, no discussion of the merits of a scientific advance would be complete without ye olde “a great experiment leaves you with more questions than answers.” I think it can be safely said that from this point, entire lines of inquiry sprang up which are still interesting 35 years later. As alluded above, the question about whether they indeed functioned as a cascade arose, there were equally appealing possibilities that must of them functioned at the transcriptional level (as proved to be the case), or whether they influenced cellular decisions at the translational or post-translational levels. And some answers were not apparent at all. Arbitrarily consider even-skipped, a gene whose function is required in the even-numbered, but not the odd-numbered segments. Sounds easy… if an embryo can count its own segments and divide by two. Much harder when you realize that each segment by definition is its own ‘place’, thus begging the question “how do you make a rule of expression/activity that applies in each and every even-numbered segment, but in no odd-numbered segment? The answer is a tad inelegant, but is dramatically demonstrated in the work of Small et al. above–for each segment in which expression is required, a separate chunk of regulatory DNA exists. That chunk of DNA constitutes a regulatory ‘equation’, made up of positive and negative elements that combine to bring about transcription of the nearby even-skipped coding sequence in one of the required segments.
Overall, then, a bold and insightful piece of thinking undergirded by a huge amount of work–made manageable by the insightful application of the powers of a uniquely appropriate system.