 Cell biology can be a challenging aspect of Introductory Biology. It’s visually fascinating, the techniques are now incredibly diverse and powerful… but the usual issue arises: what are the concepts that we ought to be teaching? The topics include organelles, transport, membranes, compartmentalization… but why? I think that protein trafficking draws together many of these fundamental facts in the context of engaging students in questions and wonder. I also propose a framework for turning a potentially dry listing of facts and names-of-parts into a journey of exploration where students ‘accidentally’ learn techniques, organelle roles, and scientific community.
Cell biology can be a challenging aspect of Introductory Biology. It’s visually fascinating, the techniques are now incredibly diverse and powerful… but the usual issue arises: what are the concepts that we ought to be teaching? The topics include organelles, transport, membranes, compartmentalization… but why? I think that protein trafficking draws together many of these fundamental facts in the context of engaging students in questions and wonder. I also propose a framework for turning a potentially dry listing of facts and names-of-parts into a journey of exploration where students ‘accidentally’ learn techniques, organelle roles, and scientific community.
I confess to being challenged by teaching protein trafficking in the past, despite finding the topic fascinating. There are many critical concepts in play: compartmentalization, the origins and maintenance of organization (in my view, one of the hallmarks of life), the use of lock-and-key protein:protein interaction to derive directional flow and order from random events. I have found that the whole process can be fruitfully compared to a post office, with zip codes attached to each protein (though it’s worth noting that this may begin to fail as snail mail dies out and students no longer have any idea about how e-mail reaches the correct destination!). I have learned that many of us have independently come to embrace this analogy.
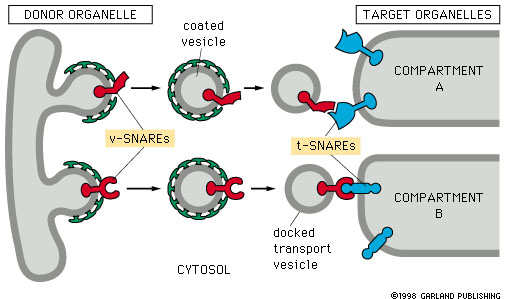
But… the details, while housing the devil, can be uninteresting. Each protein’s ‘zip code’ is a string of amino acids. While it’s fun to lead students to this conclusion (in my class, we have discussed specific interactions by this time, and the concept of making the addressing information out of the protein itself is an accessible one; from there, the idea of encoding the address at one or the other end of the protein flows easily). But how do we make this concrete to students? I show them a list of consensus signal sequences… but don’t make them memorize them, because they’re likely accidents of history, and unlikely to provide illumination.
First: the wonder of protein trafficking/targeting
In my opinion, teaching cannot succeed without first sparking interest. So, what’s interesting about a bunch of proteins going to a bunch of membrane ‘baggies’? My opinion here is the same as in other places: it’s a process that works without an overseer. We sort things because we are larger than they are, generally use visual assessment to categorize, and then manually place items to locations we also visually characterize. It’s fascinating (to me 🙂 ) that not one of these methods is available to a cell. As with distributing chromosomes, as with protein folding, as with the dance of translation and as with so many other events, the wonder is that all this stuff goes on with no one directing traffic, and no hope of the outside perspective required to do so. So I try to challenge students: “How is this even POSSIBLE?” Once we get them asking questions, I think we have an audience.
Second: the outline
So… how to avoid providing students with long lists of what proteins go to what compartments, what processes go on therein, how did we find these things out, and how do cell biologists work in general? I should pause to say that while I think that the designators are breaking down (Cell Biology, Biochemistry, Genetics, Molecular Biology–anybody can pick up anyone’s tools, and generally ought to!), I think a cell biologist is in general a watcher, a biochemist a divider and conqueror, a geneticist a breaker and re-maker of things, and a molecular biologist a subverter and re-director of cellular machines. I propose incorporating tools-of-watching (GFP, microscopy, compartment-specific stains) with those from other arenas (deletion mapping, protein fusions) and binding it all up as a whole-class exercise. While the project has taken shape in my mind, I’ll confess it remains one of those Next Big Things.
The elements that I think can/should be incorporated into an exercise/exposition on protein trafficking include
- the purpose : compartmentalization is the brilliance of eukaryotes. I tell students it’s like separating your bathroom, library and kitchen. They pretty much get that 🙂
- the implications #1 : compartments need boundaries (and what better way to separate water-filled domains than with a water-excluding ‘baggie’: a hydrophobic membrane?)
- the implications #2 : for the compartments to act differently, they must be filled with the actors that bring about special behavior
- the implications #3 : how do the required actors (proteins) end up where they ought to be, and not where they oughtn’t?
- Techniques #1: Molecular biology. Determining leader/tag sequences and grasping the concept of ‘consensus sequences’
- Techniques #2: GFP tagging, protein fusions, deletion mapping
- Techniques #3: Assigning roles to compartments (a combination of specific indicators, such as redox dyes, and biological behaviors, such as ‘following’ engulfed particles through the cell)
- Exposition: a discussion of the details-of-implementation that are unlikely to be deduced from first principles even by engaged students. Hopefully, by the time we get here, they’re eager to understand more!
Third: the delivery
Disclaimer: This is a mental work in progress (last night yielded the insight about providing students with ‘destination’ and ‘leader sequence’ tags) so this section may change over time, and will include nascent rather than polished ideas! I’m going to present the concepts linked to their delivery as I currently plan it; of course ‘someday’ a giant classroom exercise using preceptors could be replaced by computers handling the graphics, dispensation of information and tasks, and perhaps even coordinating a joint approach.
The game is afoot: getting the task started
Pair up your students (just because two heads are generally better than one, and always less frightening!). I’ll outline this for a class in the 25-30 range, or a subdivision of a larger one. Otherwise, things will just get too unwieldy. To begin, student is given just a strip of paper with some letters on it. We’re likely far enough along in the semester that they’ll recognize them as protein sequences:
Pro-Ala-Leu-His-His-Trp
Leu-Ala-Leu-His-His-Trp
Lys-Lys-Glu-Gly-Lys-Arg
Theirs won’t be colored; indeed, they’ll have no distinguishing characteristics at all. In a perfect world, these would actually be peptides, perhaps shown in flat form with charges indicated (since it is ‘feel’ rather than alphabet that causes all molecular interactions/recognitions).
Now the fun begins! Instructions to the class: “Wander among your classmates and gather with those most ‘like’ you.” You, of course, will have pre-chosen your sequences so that some are identical, and those in different groupings all diverge greatly, while those intended to cluster only differ trivially, as the two green selections above.
This, unfortunately, introduces the first design challenge and opportunity for conceptual confusion. Protein sorting is NOT done on the basis of ‘like gathers with like’; I’m wrestling with the idea of getting students to come up with the idea of a consensus sequence, something that is important in understanding promoters, cell sorting, protein-protein interaction, recognizing signals in genomes… There is an alternative that would also suffice, and might be better: once the students each have ‘their’ sequence, your preceptors arise and hold up big signposts. Your instructions: wander around the room until you find a sign matching your paper! This better captures actual cellular events. If a clever way could be found to embed the idea of complementarity then you could ice the cake: “Wander around the room until you find a ‘docking station’ that matches up with your protein structure.” This would directly reveal the underlying ’cause’ of consensus sequences: they are performing a role in interaction, and the interaction is generally ‘over-specified’ (in terms of binding energy/information content).
Have the students compare their sequences and they will see that they have ‘self sorted’ to different ‘compartments’, despite not knowing where they were ‘meant to go’ and not all following the same path or in a group together–much like proteins sort. The secret is indeed the conceptual basis of the process: individuals proteins contain peptide ‘tags’ that allow them to ‘latch on’ to destinations. Caveat: yes, this actually leaves out an entire layer–proteins are tagged, the tags interact with binding sites on the INSIDE of vesicles-to-be, and the ‘docking proteins’ (v-SNAREs) on vesicles span membranes and present binding shapes externally that will bind to surface proteins on target compartments (t-SNAREs). Depending on how exotic you are feeling, you could mimic this. With 300 students, an auditorium, and an afternoon, one could run quite the cell-sorting extravaganza, with ‘bus/vesicle-loads’ of students shuffling off to their respective compartments :-).
Where am I, and what’s going on here?
Another goal of a protein trafficking exercise is teaching students about the roles that individual compartments play in the life of the cell, the organization that makes it all happen, and the tools cell biologists use to investigate them. These will be assaulted in phase II, where students learn about the ‘compartment’ to which they have sorted themselves.
The basic principle here is going to be “How do we get students to think about what goes on in different compartments, what the machines/conditions are that fit the compartment to that role, and what tools can we provide that makes this discovery-based?” Some examples to start with: mitochondria are redox factories; reagents that change color in the presence of high-energy electron sources will flare up in a mitochondrion. They also have distinctive double-membrane structures with a maximization of surface are, so the inner membrane is folded and convoluted. Molecular Probes has created a number of tools to display their dyes, and which could be used to create stills; a page of dyes is here; a webpage simulator which used to do beautiful, wonderful things is here (but not currently working for me), but perhaps because they’ve shifted their attention to this glorious iPad app. Failing any of these options, ye Google Image search is a rich source of the wonderful things folks create.
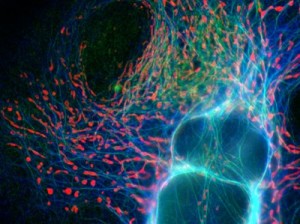
Of course, the things you tell students about ‘their’ compartment can be directly driven by the concepts you want them to take away. Do you want a structural description, as if they woke up in a cave of wonders? If in a newly formed endosome, do they shiver as the pH drops and feel fear as marauding proteases are let loose? Do they see flu virus particles, apparently captured, proceed to harpoon their way out of the cell?
In the Golgi, are they ‘decorated’ with sugars, perhaps each student consulting the back of their original tag (piece of paper with protein sequence) and getting hooked up with preceptor-glycosylases who provide them with sugar ‘wristbands’ corresponding to their further sequence information. And pointing out to them that their travels are not over; they must consult further information yet (or be handed it) and be on their way again.
Active experimentation
This is where I’m getting foggy on “What to leave in/what to leave out” (Bob Seger, “Against the Wind”). Unfortunately, one cannot both start them off with just the targeting tag and proceed to have them experimentally determine which region of the protein is responsible for targeting by using a combination of GFP tagging and deletion analysis. Perhaps this belongs in the follow-up lecture on “how did we come to know these things”
Other pieces that… belong here?
I think the molecular details belong in the follow-on lecture as well, but the hope is that having ‘lived the life’, students are ready to comprehend the details, and see how order arises from chaos.
I think it would be interesting to try to work in Signal Recognition Particle, stopping translation until an ‘endoplasmic reticulum entry port’ is encountered, etc. But it’s not clear how to do that mechanically. My bias would be to tell that as a story as a preamble to each student embarking on their own journey. A cute way to make the point would be to send the students home from the previous lecture with a ‘ticket’ to the classroom, which happens to be the SRP signal sequence. They are required to present this outside the classroom to monitors, and upon doing so, gain entry to the ‘membrane system’ of the lecture hall. [Yes, I’m aware fully 30% would forget them–maybe we sell ’em outside for $1.50?]. This would hopefully have students curious for 15 minutes of lecture transitioning them from existing knowledge about translation and preparing them to enter this next stage of their understanding.
Appendix: Yeah, we really do know that stuff
The consensus signals have been figured out over the years. I think asking students to memorize them is deeply pointless, but there’s no reason not to use the actual signals for realism in the exercise, and to let them look over what a signal looks like…in print and linearly, which of course provides limited insight to what one really looks like! Source: Wikipedia
Take me to the E.R.–stat!
H2N-Met-Met-Ser-Phe-Val-Ser-Leu-Leu-Leu-Val-Gly-Ile-Leu-Phe-Trp-Ala-Thr-Glu-Ala-Glu-Gln-Leu-Thr-Ly-Cys-Glu-Val-Phe-Gln
Keep me in E.R.; return me if I stray
(KDEL: Lys-Asp-Glu-Leu) [otherwise, allow progression through the system]
I’d like to go to the nucleus please:
Pro-Pro-Lys-Lys-Arg-Lys-Val
Mitochondrion, if you would:
H2N-Met-Leu-Ser-Leu-Arg-Gln-Ser-Ile-Arg-Phe-Phe-Lys-Pro-Ala-Thr-Arg-Thr-Leu-Cys-Ser-Ser-Arg-Tyr-Leu-Leu
Peroxisome, and step on it!
Ser-Lys-Leu-COOH or -Arg-Leu-X-X-X-His-Leu
I’d like the lysosome:
mannose-6-phosphate