The main point in teaching with themes is to give students a meaningful sense of familiarity so that their travels from topic to topic are visits with friends and known quantities rather than onslaughts of fresh vocabulary and new un-anchored idea-pieces. One triplet that I have enjoyed using in introductory biology is the set combining 1) EF-Tu and its role in promoting accuracy in translation, 2) the GTPase timer in tubular that gives rise to the grow-catastrophe-disassemble cycle in microtubules, and 3) the ‘self shut-off’ timer features of many G-protein coupled receptors. These are evolutionarily related machines and each applies the concept of ‘timing’ to a different biological circumstance. Thus, besides providing three examples of how biological machines solve problems, this theme ties in to previous ones on ATPase (GTPase mechanism is identical; the attached base has nothing to do with catalysis) and evolution through duplication and modification.
‘Thinking like a molecule’ is a challenge for students because many of their intuitions from the macro- world and their human senses don’t transfer to the molecular world. For example, in proofreading of nucleotide additions, we would simply “make a mental note” that we need to ‘be careful’. And if we wanted to know if a pairing was correct, we would ‘look at it’ and see if it matched our mental dictionary of pairings. But molecules don’t have minds or memories, and the only sense at the molecular level is touch. So the question of how molecules bring about behaviors such as “be careful” and “wait a while” and “make sure everything fits correctly” are not only fascinating, they are essential to a meaningful understanding of how cells do their business–how life works.
One common solution that cells have ‘found’ is to measure time and indirectly measure things based on the passage of time and its consequences. For example, a ‘good’ basepair (one forming 2-3 unstrained hydrogen bonds) will last longer against the forces of Brownian Motion than one forming fewer bonds or bonds accompanied by stresses (such as charge-opposition). A well-matched codon-anticodon interaction forms with similar speed and facility to a poorly matched (only 2 of 3 bases paired) one… buts falls apart 1000 times faster. So if the cell could hardwire in the act of “wait-and-see”, then it could achieve accuracy by incorporating the concept of “good things last; bad ones are ephemeral”.
The second prong of the key elements here is what it means to be an enzyme. The classic definition suits us: it’s a machine that speeds up a process that is favorable, but to which there is some form of impediment. Normally, we think of the best enzyme as one that does its job as quickly as possible, but there are many cases where that’s not the case. Consider: given an enzyme with a non-limiting supply of substrates, the design of the enzyme will determine the reaction rate. We can express this concept either as number of reactions per unit time, or amount of time per single reaction. It’s this latter that interests us here. Given an enzyme of ‘intermediate’ ability, let’s say that on average it executes its reaction in 100 time units. We can therefore also think of this machine as a timer! The more efficient the enzyme is, the shorter time lapsed between grabbing substrate and completing the enzymatic chemistry. Since ‘being an enzyme’ is nothing more than creating an environment consisting of amino acid sidechains and space in 3 dimensions, a given machine can be made more or less efficient–it’s timeperiod can be ‘tuned’ to be faster or slower!
How does this concept apply to our three biological timers?
EF-Tu as a biological timer in translational accuracy:
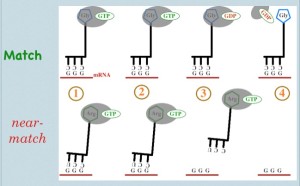
Tutorial video. [Note: EF-Tu is the bacterial version of eukaryotic elongation factor eEF-1]. EF-Tu ‘chaperones’ tRNA-amino acid couples. Functionally, it blocks the amino acid in such a way that it’s amino-terminus is inaccessible. Recall that when a tRNA is added to the protein chain during translation, the actual event is that the new amino acid takes the existing chain from the previous amino acid. While almost never portrayed in textbooks, this step is impossible while EF-Tu is present… and EF-Tu is present on all circulating tRNA-aa pairs. So what induces EF-Tu to leave? Briefly, upon entry into the ribosome and an apparent ‘match’ between tRNA anticodon and mRNA codon, the ribosome induces EF-Tu to become a decent enzyme (recall from above that ‘good’ codon-anticodon matches and ‘wrong’ ones form with similar facility; the key is how fast the wrong ones fall apart). From this point, the EF-Tu GTPase is ‘worrying’ at the gamma phosphate of GTP, attempting hydrolysis. Is it the greatest GTPase ever? No. This process takes time–an amount of time evolutionarily optimized to split the difference between the (relatively) long time a correct codon-anticodon match will last, and the (relatively) short time a 2-of-three codon-anticodon will match.
There are two possible outcomes: 1) EF-Tu hydrolyzes GTP successfully, GDP remains and phosphate leaves => conformational change of EF-Tu => EF-Tu loses affinity for tRNA-aa => EF-Tu dissociates => amino group of new amino acid is exposed => ribosome can do its thing, and the new amino acid grabs the existing chain or 2) Brownian motion drives dissociation of codon from anticodon before GTP hydrolysis => tRNA loses ‘grip’ on codon => tRNA + amino acid + EF-Tu exit the binding site => poor-fitting tRNA leaves without erroneously joining the elongating peptide chain.
Tubulin and microtubules biological timers trigger ‘catastrophes’:
One of my favorite processes in cell biology is ‘chromosome catching.’ Dozens of chromosomes are scattered about, each with a tiny target (centromere). The cell has limited time to achieve this, doesn’t ‘know’ where they are, and must proceed ‘blind.’ Formally, it’s an impossible task–to search the necessary volume, the cell would have to create far more microtubules than it has resources to build. What to do? Briefly, follow the strategy of the fly-fisherman–with a large volume to search and a small hook, repeatedly ‘cast’ the line into the potential area and drag it through (in the case of microtubules, we ‘push’ the line instead of pulling it). If a successful collision occurs, that chromosome is ‘caught’; make the line fast and wait for the rest to be landed.
I’ve over simplified here (I hope to add the entire lecture series I do under Lecture Modules ‘someday’), but that’s the essence. This leaves us with another tricky problem. Microtubules aren’t ‘things’; they’re long polymers of individual tubulin dimers. How do we achieve two kinds of behavior–GROW GROW GROW GROW followed by collapse and disassemble–when the components don’t know what the population is doing? Again, oversimplifying, the essence is a combination of the difference between GTP- and GDP- bound tubilin dimers combined with the fact that new units are added to one end of a microtubule and conceptually constitute a ‘cap’
Features distinctive to GTP-bound units include
- this is the state of free tubulin in the cytoplasm
- only hydrolyze to GDP after addition to end of growing microtubule
- relatively stable within and and end of microtubule
- are good ‘building blocks’ for continued microtubule growth
Features distinctive to GDP-bound units include
- likely cause unraveling of end of microtubule
- awkward for further addition/growth of microtubule
- likely to dissociate from end of microtubule
Thus in the life of a microtubule, a constant race-agaist-time is going on. If additional tubulin subunits are added relatively quickly, the microtubule terminus consists of a ‘cap’ of GTP-bearing tubulin units. These support continued growth, and by the time they ‘burn’ their GTP down to GDP, new GTP-bearing units have been added, and growth continues. Old, GDP-bearing tubulin units are trapped by the structure of the tubule and cannot ‘express’ their tendency to unravel and dissociate. However, if by chance there is a slow period of tubulin addition or there is a chance-driven high frequency of GTP-to-GDP conversion at the tip, everything changes. The tip begins to fall apart, new additions are rare and hard to bring back to stable structure, tubulin units fall off… revealing still-older units behind them. These, of course, have had tons of time and likely all converted to GDP-state. So they, too unravel and the ‘catastrophe’ (that’s the technical term) continues until the tubule shrinks to nothing.
Where’s the timer? The essence of this whole behavior is the ‘tuning’ of the conversion of the tubulin dimers in going from GTP-bound to GDP-bound. If the GTP-state is ‘too stable’, then the tubule will try to grow ‘forever’, limited only by the supply of tubulin in the cell. If too unstable, then tubules will disintegrate before much growth is achieved, and most of the space where chromosomes could be hiding will never be ‘explored’. If tubulin had only a single state, they’d likely muddle about at an intermediate length, neither growing nor shrinking much–so the cell would explore a few locations REALLY, REALLY well–but not achieve its goal of finding ALL the chromosomes.
The idea of adjusting kinetic parameters to bring about an emergent behavior such as ‘microtubule fishing’ can be challenging; thinkBio ‘Tubule‘ allows students to explore chromosome capture by adjusting basic parameters (ATPase, tubulin add/fall off rate) and observing a simulation of chromosome capture.
As one always finds with biology, there is further regulation on the innate ‘setting’ of microtubules; there are a whole host of molecules whose presence or activating ‘tweak’ the GTPase rate to be slower (increase the ‘growth’ tendency of the affected tubule) or faster (accelerate catastrophe and disassembly)
Shutting down a signal: GPCR off switches are biological timers
(G-protein coupled receptors; technical but complete Wiki article). Signaling only works if the signal STOPS sometimes; otherwise, the first thing you saw when you opened your eyes would be the only thing you saw… forever. This can get expensive (and does)–not only must there be mechanisms for turning ‘on’ a signal; the same signal must be shut down, and at all levels lest it just fire up again. One elegant way this is achieved in GPRC’s utilizes the same timing mechanism covered above. I’m going to skip 95% of the signaling pathway here, because this post is growing long. Briefly, when the receptor itself (trans-membrane protein; often these receive signals in the form of small molecules on the ‘outside’, though opsin, for example, uses an internally held molecule of retinal to generate the signaling state) is ‘tweaked’, the resulting conformational change is structurally transmitted to the inside of the cell.
The consequences of change on the membrane side are aways the same: the activated GPCR begins converting all the matched protein trimers with which it associates to active form. These are the G-proteins, which consist of alpha, beta, and gamma subunits. When packaged as a party of three, they’re inactive and the alpha subunit bears a molecule of GDP. Activated receptors recognize ‘their’ specific G-protein(s) and catalyze the exchange of GTP molecules for GDP When the alpha subunit gets its GTP, it dissociates from the others, and it’s off to the races! (as always, with exceptions and variants; in yeast, it’s often the beta-gamma unit that does the signaling). GTP-bound alpha subunits activate targets for which they have high affinity and the signal is ‘live’ inside the cell.
How to put the genie back in the bottle? Recall that the signaling state was brought about by swapping in a GTP molecule. And there is little in the cell easier than busting up a molecule of ATP/GTP… especially when there is no associated work to be done! As with the other two, given that one could conceptually make a highly efficient GTPase (a timer that fires very quickly), one can readily imagine ‘tuning’ a specific GTPase for whatever level of activity chosen–and the worse the enzyme (i.e., longer it will take, on average, to execute GTP hydrolysis) the ‘louder’ the launched signal will be before it winds down. And since the alpha subunit of the G-protein is the signal, no spare parts are required! That said, many systems further fiddle with the system by adding other proteins that speed up the innate hydrolysis or regulate it.
Overall, then, we see the same physical component (a GTP-binding/hydrolysis domain in which the removal of that last phosphate of GTP triggers a conformational change) acting as biological timers. In translation, the ability of a tRNA anticodon::mRNA codon match to ‘wait a while’ is a measure of its quality and thus accuracy; in the growth of microtubules, the timer in the terminal units creates a competition to add new subunits or fall apart, generating cycles of grow-grow-grow that convert wholeheartedly to disintegrate, and in G-protein coupled receptors, the constant attempts of the alpha subunit to hydrolyze GTP generate a self-terminating aspect to signals once they are initiated.
IN the classroom, the first instance is very hard. Students have never heard of EF-Tu, and the cellular ‘way’ is foreign to them. Textbooks tend not to entertain the concept that a MIS-matched tRNA could ‘meet’ the mRNA, and it’s a challenge for them to find a way to distinguish good from bad fits or to build a machine that implements those ideas. So in my hands, this is a lecture accompanied by pauses for thinking questions. At the end, though, students that master these ideas have a real idea of how the cell does things, and are in a great position to apply this conceptual understanding to the two very different appearing cases that follow. The net is that they have begun to understand how cells truly work–proteins with simple, comprehensible, mechanical behaviors act to bring about complex outcomes at the heart of life itself.
Addendum: Cruising through some of my old collections, I just found this paper, which is really just more of the same–here, the timing function is involved in ordering events of membrane/vesicle fusion during intracellular transport. Also re-visits a clever scheme used elsewhere to make convert the GTPase activity to XTPase (xanthosine triphosphatase) by changing a single amino acid position in the protein to match a single basepairing difference between xanthosine and guanine.